Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Nasir Arefinia1, Hamid Reza Mollaei*1, Seyed Alimohammad Arabzadeh1 and Ali Akbar Haghdoost2
Received: September 25, 2018; Published: September 28, 2018
*Corresponding author: Hamid Reza Mollaei, Assistant Professor, Department of Medical microbiology, Kerman University of Medical Sciences, Kerman, Iran
DOI: 10.26717/BJSTR.2018.09.001805
Background: To evaluate frequency of Cytomegalo virus resistant to ganciclovir a cross-sectional study was designed.
Methods: In this study 96 samples from patients infected with HIV, 41 males and 55 Females with Mean ± SD years old age 52.6 ± 6.9 and 42.6 ± 9.1 respectively were collected. The both buffy coat and plasma were tested using Real Time PCR for detection of CMV, because the CMV virus latent in Peripheral Blood Mono Nuclear cells (PBMNs).
Results: Out of 96 patients, twenty-two (22.9%) buffy coat samples, and seven plasma samples (7.3%) were positive for CMV. A High-Resolution Melting (HRM) curve assay was performed for detection of CMV resistant to ganciclovir in UL97 region. Out of 22 CMV positive samples from buffy coat, there were five samples resistance to ganciclovir (22.72%), three M460V, one M460I and one mix genotype of M460V/I. Out of seven positive samples from plasma four samples (57.14%) positive for resistance to ganciclovir.
Conclusion: Mutation in UL97 and UL54 regions can be resistance of CMV to ganciclovir. High-level resistance to ganciclovir often is associated with cross resistance to other polymerase inhibitors such as cidofovir. Most point mutations lead to resistance to treatment with ganciclovir was done at 460,594 and 595 amino acid location in UL97 gene. Today, we are witnessing the emergence of drug resistance in viruses. This causes diseases that previous therapies do not affect and cause mortality in humans. Therefore, the evaluation of drug resistance in viruses is of importance and can provide a better approach to physicians in treating their patients.
Keywords: Cytomegalovirus; Drug resistance; UL97; Ganciclovir; HRM
Abbreviations: CMV: Cytomegalovirus; CNS: Central Nervous System; GI: Gastrointestinal; dGTP: Deoxyguanosine Triphosphate; ART: Antiretroviral Therapy
The cytomegalovirus (CMV) is one of the most causes opportunistic infections in people with AIDS and the CMV most commonly encountered retinitis in 80% of patients infected with HIV Adachi et al. [1]. CMV is a major cause of morbidity and mortality in patients with HIV in the United States. CMV infection causes disease in several organ systems, including the central nervous system (CNS) (chorioretinitis, encephalitis, polyradiculopathy, myelopathy) and the gastrointestinal (GI) tract (oral ulcers, esophagitis, hepatitis, colitis, intestinal perforation), as well as life-threatening adrenalitis and pneumonitis ArellanoGalindo et al. [2-4]. Epidemiologic studies indicated that half of HIV patients which are affected by CMV; have symptoms including chorioretinitis, esophagitis, colitis, pneumonia, and central nervous system disease Gianella et al. [5]. Diagnosis and detection of CMV disease can be based on clinical evaluation, such as CMV retinitis and serological or molecular assays. Culture of CMV from blood, urine, or even biopsy of tissue is very time consuming Leila et al. [6]. Detection of CMV inclusions, antigens, antibodies or nucleic acids are better and easier than culture methods and the use of molecular methods can be identified as quickly and accurately as possible Monavari SH et al. [7].
After identifying people infected with the CMV, the most important treatment is correct and timely treatment Stoeva et al. [8]. One of the most commonly used medications in the treatment of CMV is the ganciclovir. Ganciclovir is a synthetic analogue of 2′-deoxy-guanosine. It is first phosphorylated to ganciclovir monophosphate by a viral kinase encoded by the CMV gene UL97 during infection Vanpouille et al. [9]. Subsequently, cellular kinases catalyze the formation of ganciclovir diphosphate and ganciclovir triphosphate. Ganciclovir triphosphate is a competitive inhibitor of deoxyguanosine triphosphate (dGTP) incorporation into DNA and preferentially inhibits viral DNA polymerases more than cellular DNA polymerases Bachmann et al. [10]. In addition, ganciclovir triphosphate serves as a poor substrate for chain elongation, thereby disrupting viral DNA synthesis by a second route BischoffJung et al. [11]. Drug resistance occurs in patients receiving longterm anti-CMV drugs. Rates of resistance approximately 25% per person-year were reported and similar for ganciclovir, foscarnet, and cidofovir. In the antiretroviral therapy (ART), the rate of resistance appears to be lower (approximately 5% per personyear) Bruminhent et al. [12]. Low-level resistance to ganciclovir occurs through mutations in the CMV UL97 (phosphotransferase) gene, and high-level resistance to ganciclovir typically occurs in both the CMV UL97 and UL54 (DNA polymerase) genes Chen et al. [13].
Resistance to foscarnet or cidofovir occurs in the CMV UL54 gene mutation. High-level resistance to ganciclovir often is associated with cross resistance to cidofovir and occasionally to foscarnet Chevillotte et al. [14]. Sequencing of the UL97 gene from PCR-amplified specimens can be accomplished in less than 48 hours, correlates well with conventional drug susceptibility testing and clinical outcomes, and therefore has clinical utility for patients in whom therapy has failed Chou et al. [15]. Conventional methods of culture and susceptibility testing and viral sequencing often are not available in clinical laboratories because they are too time-consuming or costly. Peripheral blood CMV viral load measurements have poor positive predictive value for treatment failure Chou et al. [16]. Patients with high-level ganciclovirresistant isolates will require a switch to alternative therapy. Most point mutations at 460,594 and 595 amino acid location in UL97 gene, lead to resistance to treatment with ganciclovir Daikoku et al. [17]. In HIV patients that infected with CMV, treatment with ganciclovir should be continued in the form of oral administration 2-4 weeks after treatment to injectable form. This can prevent of the development resistance to ganciclovir, and continuing treatment is the most important and reduces incidence of mutation Garrigue et al. [18]. Aim of this study, evaluation frequency of CMV infection and detection of resistant strains of ganciclovir in HIV patients by analyzing the UL97 gene of CMV positive samples.
In this Cross-sectional study, 96 patients (38.66±11.29 Years old age) participated with the HIV who referred to the Center for Behavioral Illnesses in Kerman province. After receiving informed consent and explanation of the project, blood samples were taken from these patients. Their blood samples were sent to appropriate laboratories for monthly tests including HIV load, CD4 count, SGOT, SGPT, and Urea. Of the 96 patients in this study, 55 were male (57.3) and 41 were female (42.7%). Age group distribution each of sex was demonstrated in Figure 1. The highest age group in men and women, the age group between 20 and 40 years old (28.13%) and 21.88% respectively. All patients had been treated with antiretroviral drugs for more than three months. Exclusion criteria included a coexisting severe illness, organ or bone marrow transplantation, recent treatment with systemic corticosteroids, and chemotherapeutic agents.
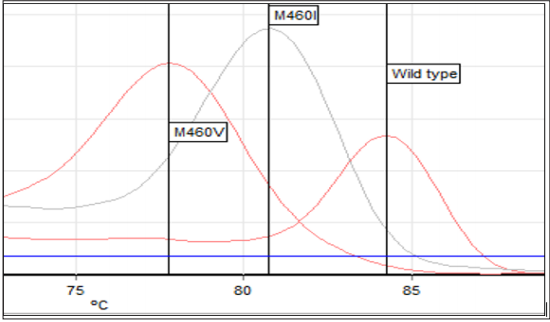
Figure 1: Specific Melting temperature of UL97 wild type and mutation strains (M460V, M460I). Melting temperature of wild type: 84.2°C; M460V:77.8°C; M460I:80.8°C.
About Five ml of peripheral blood were collected from each patient into EDTA-containing vacutainer tubes. Plasma and Buffy coat were separated and stored at -70°C. CMV DNA, HIV RNA were extracted from 200 μL of plasma and Buffy coat with High Pure Viral Nucleic Acid kit (Roche Diagnostics GmbH, Mannheim, Germany). Extracted DNA/RNA pellets were resuspended in 100μL of prewarmed Elution buffer and stored at -20°C until use. For Quantification of CMV DNA, HIV RNA we use Real Time CMV, HIV kit (ILS, Russia). Quantitative determination of the amplified products was done with the Rotor Gene Q, 5-Plex (Qiagen, Germany).
We present a new HRM(High Resolution Melting) PCR assay using specific UL97 primer with melting point analysis for characterization of the relevant UL97 codon 460 .The binding of the primers leads to a specific melting point for the wild-type sequence. In mutant strains, there is the mismatch between two primers and the mutant target sequence results decreasing the melting temperature. The following program was used for cycling: initial denaturation, enzyme activation at 95°C for 10min; denaturation step at 95°C for 20 s; annealing and elongation steps at 60°C for 40 s, repeat for 40 cycles; and melting curve analysis at start in 60°C up to 95°C for 10 s, with a slope of 0.1 (°C/s). For increase of sensitivity of test we use Accu Melt HRM SuperMix (Quantabiosciences, USA).
HRMA with unlabeled probe proves to be a sensitive and cost-effective genotyping method for the detection of M460V/I mutations. The product size of UL97 PCR was 470 bp. Primers were design using Beacon designer software (Version 8, Primer Biosoft, and USA). Sequences of primers were UL97F (141693– 141712) Forward primer CTGCTGCACAACGTCAAGGT, UL97R (142131–142153) Reverse primer CCCAGCGCCGACAGCTCCGACAT. Quantitative determination of the amplified products was done with the Rotor Gene Q (Qiagen, Germany). The wild type strains of UL97 had a specific melting point of 84.2°C, while the point mutations in codon 460 reduced the specific melting point to M460V:77.8°C and M460I:80.8°C (Figure 1). The difference between melting points for the wild type strains and the UL97 mutant strain (M460I, M460V) was 6.4°C for M460V and 3.4°C for M460I.
Chi square and Fisher’s exact Tests were used to analyze the data obtained by SPSS 11.5 software (SPSS Inc, Chicago; USA). The differences or association with p<0.05 were considered statistically significant.
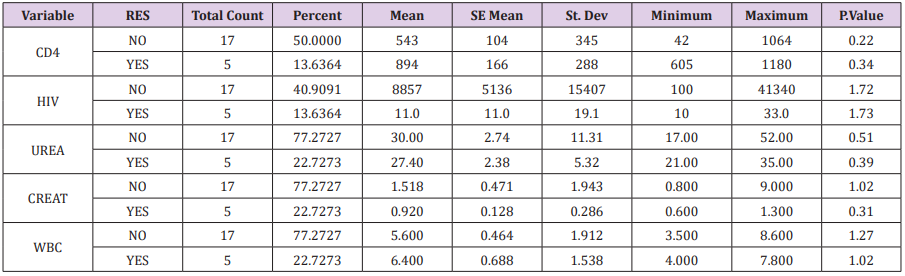
In this cross-sectional study, in total of 96 patients with an average age of 38.6 ± 11.2 , which were 55 females (57.3%) with an average age of 42.6 ± 9.1 and 41 males (42.7%) with an average age of 52.6 ± 6.9. Table 1 was shown Mean±SD and other parameters in 96 patients. Various parameters such as white blood cell count, liver tests, and renal function tests were performed annually on all of samples, the results of which are summarized in Table 2. After performing the Real time PCR test on plasma and Buffy coat samples, Seven (7.3%) and 22(22.9%) samples were positive respectively for the presence of CMV DNA. From 22 positive samples, 14 samples were female (33.33±12.94) and 8 samples were male (41.71±6.32) years old age. The highest frequency of CMV Positive was shown in the age group of 20-40 years (10.42%) and 40 to 50 years (8.33%) (Figure 2). After HRM test for detection of mutation in UL97, Out of 22 CMV positive samples from buffy coat, there were five samples resistance to ganciclovir (22.72%). From positive resistance samples there were 2 male (40%) and 3 females (60%)(Table 3). The highest drug resistance to ganciclovir was also in the age group of 20 to 40 years (2.08%) in both male (35±2.83) and female (39.3±19.9). In Table 4 significant relation between different parameters in CMV positive patients and, CMV resistance was shown .There was no significant relation between CD4 count, HIV load, Urea, Creatinine, SGOT, SGPT and CMV resistance in HIV patients.
Table 4: Statistic characteristic in 22 positive samples and five samples resistance to ganciclovir.
Human cytomegalovirus (CMV) results in serious complications in immunocompromised patients. Tissue-invasive CMV disease, such as pneumonia and hepatitis, is responsible for high morbidity and mortality rates in transplant patients Kim et al. [16,17]. The use of prophylactic anti-CMV therapy beginning early post transplant has reduced the incidence of CMV-associated morbidity and mortality. However, with the increased administration of anti-CMV drugs, there has been an increased emergence of drug-resistant CMV Lopez-Aladid et al. [18]. The incidence of drug resistance varies depending on the organ transplanted and the immune suppressive regimen, but it is commonly seen in 5 to 10% of transplant patients who are CMV seronegative receiving a CMV-seropositive organ Piret et al. [19]. Studies have shown that viral treatment of CMV by ganciclovir is associated with high drug resistance to HIV patients. Because in this treatment, many anti-polymerase inhibitors (HAARTs) were used and virus to escape from the treatment by creating a mutation in the UL97 and UL54 genes, causes resistance to the ganciclovir drug Vanpouille [20].
Studies have also shown that 38% of strains resistance to ganciclovir isolated in urine of patients which receiving and treated more than 3 months with ganciclovir Rolling et al. [21]. Two factors of excessive consumption of drugs and the duration of drug use are important in the development of resistant strains. The mutation in the UL97 gene can lead to a change the Methionine to the isoleucine and the formation of M460I; change the Methionine to Valine and the formation of M460V and change the Methionine Threonine and the formation of M460T, M460V and M460T mutations have the highest and lowest incidence, respectively Zeng et al. [22]. Also, the different mutation has been identified in other genes such as UL54, which plays a role in polymerase Schnepf et al. [23]. In the present study, 96 patients treated with ganciclovir for an AIDS-related CMV disease were followed up over 2 year by a CMV Real Time PCR assay. The CMV isolates were tested for ganciclovir susceptibility for the UL97 gene sequence. Twenty two point seventy two percent of the CMV strains isolated from these patients were ganciclovirresistant. The results of this study are very important.
The high prevalence of CMV ganciclovir resistant strains (22.72%) can be a serious warning to physicians for treating patients with CMV positive. Molecular testing to identify mutant strains is necessary in transplant recipients for choosing a suitable treatment. Concerning associated with the natural polymorphism of viral enzymes, different mutations previously reported in the DNA polymerase Sahoo et al. [24]. In our study, M460V/I mutations were detected in five CMV isolated resistant to ganciclovir. Thus, the exact role of these mutations, natural polymorphism or compensatory mutation remains unclear and needs further investigation. Moreover, six novel mutations of unknown significance in UL97 and UL54 genes have been identified. The change V498I in UL97 phosphotransferase is located between the two conserved regions VII and IX of the protein Sharma et al. [25]. Similarly, the five novel mutations in UL54 DNA polymerase lie within nonconserved regions of the protein. In particular, 4 of them are located between domains delta-C and II (P608S, T610M, G629S) and between domains III and I (S880L), where a natural polymorphism of the enzyme Hantz et al. [26]. The last amino acid change E315D is located between the N-terminal region and domain IV. Because of their location outside the conserved domains, all these mutations are likely considered to be associated with the natural polymorphism of viral proteins, rather than antiviral resistance Alain et al. [27].
Nevertheless, further studies are required to ascertain the true nature of these novel mutations. In a study in Taiwan studied 40 clinical isolates to detect the mutations of UL97 and UL54 that might be related to resistance. The results showed that no mutation known to cause ganciclovir resistance was detected in any strain, but some polymorphisms (N685S, A688V, A885T, N898D in UL54; D605E in UL97) were frequently observed. Results suggest that resistant HCMV strains are not prevalent in Taiwan Shao et al. [28]. In a study by Smith DM and colleagues in 2016, asymptomatic replication of CMV in the early stages of HIV infection was associated with a reduction in the CD4 / CD8 ratio during the HIV treatment period Smith et al. [29]. Also, in a study by Leenasirimakul and colleagues in 2016 in Thailand showed that HIV infection with CD4 counts less than 100 cells /μl is a risk factor for cytomegalovirus infection Leenasirimakul et al. [30]. Our data is very important information about the antiviral resistance of CMV. Genotyping tests are useful in practice because they provide rapid diagnosis and can be more sensitive to phenotypic tests.
Phenotypic resistance assays may be two to four times less sensitive to detect drug resistance due to mutation and cannot detect when the mutant virus is less than 20% of the total virus population. In contrast, molecular methods, can detect genotypes created resistance inducing mutations in 10% of the viral population. Due to the fact that CMV can latent in peripheral blood mononuclear cells and has the potential of reactivation in patients with HIV, it produces clinical manifestations such as colitis, retinitis, encephalitis, and esophagitis. It is suggested that when a patient returns for periodic testing, contamination with CMV were checked. In patients with HIV and AIDS, due to immune deficiency, the antibody against CMV is low or no detectable, therefore, it is necessary to use molecular methods such as PCR to detect and identify CMV-infected patients. Also, Due to several mutations in the main genes of the CMV virus, progression patients with HIV, can be facilitates to diseases such as retinitis, encephalitis and ultimately death [31-33].
Use of Conventional PCR amplification for detection of UL97 mutation strains might be fail to detect low levels of virus, detect only the predominant type, or detect a mutation when it represents a minor population in a mixed CMV population, so, we recommended use of a sensitive and specific methods such as Real Time PCR base on TaqMan probe assay for detection of mutation strains. The relatively low prevalence of UL97 in this study may be due to the fact that in this region, the use of ganciclovir has not taken place, or mutations have taken place in other genetic regions, so it is suggested that other genetic regions such as UL54 (polymerase enzyme) should be investigated.
The author so thanks from staff of the Center of Behavioral Diseases in Kerman Province. This project has been approved and funded by the Medical Ethics Committee, the Physiology Research Center of Kerman University of Medical Sciences (Approval Cod is IR.KMU.REC.1395.860).


