Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Noboru Motohashi1, Jyothirmayi Vadapalli2, Vanam Anuradha3 and Rao Gollapudi4*
Received: May 20, 2018; Published: May 31, 2018
*Corresponding author: Rao Gollapudi, University of Kansas, Lawrence, KS 66045, USA
DOI: 10.26717/BJSTR.2018.05.001149
Heat shock protein 90 (Hsp90) is a molecular chaperone, indispensable for the stability and function of several conditionally activated or expressed signaling proteins. Hsp90 inhibitors can cause the inactivation, destabilization and eventual degradation of HSP90 client proteins by interacting predomently with a singular molecular target. Hsp90 inhibitors are unique in such a way that even though they are directed towards a specific molecular target, they simultaneously attenuate multiple signaling pathways that frequently interact to promote cancer cell survival. HSP 90 inhibitors displayed promising antitumor activity in preclinical model systems. The herbs containing alkannin (3) and shikonin (4) are used in the treatment of measles, smallpox, sores, ulcers and wounds and skin eruptions in traditional Chinese medicine (TCM). Furthermore, the present molecular docking studies of alkannin (3) and shikonin (4) suggest that alkannin (3) and shikonin (4) may inhibit cancer progressin by binding to the active site of Hsp90 comparable to Hsp90 inhibitor drug, geldanamycin (2). A detailed in vitro and in vivo studies on alkannin (3) and shikonin (4) need to be pursued to substantiate the reported the docking study observations.
Keywords: Heat shock protein 90 (Hsp90); Adenosine triphosphate (ATP); Alkannin; Shikonin; Geldanamycin; Lipinski’s rule of five; Docking studies; Lipinsky’s rule of five
Keywords: Ala: Alanine; Asn: Asparagine; Asp: Aspartic acid; Glu: Glutamate; Gly: Glycine; Ile: Isoleucine; Leu: Leucine; Lys: Lysine; Met: Methionine; Ser: Serine; Thr: Threonine; Tyr: Tyrosine; Val: Valine; ATP: Adenosine triphosphate DNA: Deoxy ribose Nucelic Acid; TCM: Traditional Chinese Medicine
Heat shock proteins (Hsp90) are molecular chaperons assisting in folding, activation and stabilization of client proteins of cells when predisposed to stressful environment. Cariogenic cells proliferate at an uncontrollable rate under stressful conditions such as hypoxia and depletion of nutrients. Heat shock proteins expression is decisive for the stability of cellular proteins, when cellular proteins are exposed to the stressful environment. As a result, Hsp90s are overexpressed in cancer cells, dissimilar to normal cells to maintain their higher rate of metabolic function for their multiplication. Thus, inhibition of Hsp90 cripples multiple client proteins, subsequently accelerating the progression of disease and maintains homeostasis of normal cells [1,2]. Hence, Hsp90 captivated considerable attention as attractive target in drug discovery to interrupt Hsp90 chaperone function [3].
Hsp90 proteins comprise of three functional domains- adenosine triphosphate (ATP)-binding, protein-binding and dimerizing domain, each playing important role in maintaining protein function. The region of protein near N-terminus has high-affinity to ATP-binding site. ATP (1) (Figure 1) binds to a sizable crevice in the side of protein which is about 15 A0 (1.5 nanometres). Hsp90 amino acid residues Ala-117, Ala-121, Gln-123, Ala-124, Gly-125, Ala-126 and Ser-129 are located in N-terminal domain active site. Here ligands adhere to prevent ATP from clamping to the active site thereby activating protein function as chaperone. Hence, HSP 90-ATPase binding region is forthwith under vehement study as it is antecedent site of drugs targeting the protein [4-7]. Several natural products were reported as inhibitors of Hsp90 by binding to C- and N- terminal domains. Geldanamycin (2) (Figure 1) inhibits Hsp90 function by binding to N-terminal domain obstructing the binding of ATP. On the other hand, novobiocin binds to C-terminal domain of Hsp90 and interrupts Hsp90 activity by impeding the binding of cochaparome CD37. Several natural products, celasterol, quinone methideterpene, gedunin, deguelin, epigallocatechin gal-lagate and gambogic acid with antitumor activity are reported as Hsp90 inhibitors where other mechanisms of their mode of action are not well understood [8]. A selective number of compounds, under clinical trials for Hsp90 inhibition, proclaimed cardio, hepato and ocular toxicity, inspiring further search for new Hsp90 inhibitors with manageable side effects.
Napthoquinones, alkannin (3) and shikonin (4) (Figure 2) are the major constituents of Alkanna tinctori and Lithospermum erythrorhizon (Family: Boraginaceae). Traditionally, they were employed as natural dye, food additive, in cosmetics preparations and folklore. Furthermore, they were wielded for treating ulcers, measles, smallpox, wounds, inflammation, sores and skin eruptions in folklore. Alkannin (3), shikonin (4) and their derivatives were reported to possess diversified variety of pharmacological activities, immuniostimulatory, analgesic, angiostatic antithrombatic, anti-bacterial, anti-viral, anti-inflammatory and anti-ulcer [9-16]. The proclaimed antiulcer and antitumor properties of alkannin (3) and shikonin (4) persuaded us to investigate possible mode of action using in Silico molecular docking studies. The contemporary molecular docking stimulation study advocated that alkannin (3) and shikonin (4) disclosed competitive inhibition with ATP (3) at ATP-binding site of N-terminal domain of Hsp90.
At exploratory stage of drug discovery, the drug likeness should be assessed accurately to reduce cost and eliminate the chance of choosing a candidate with false positive results. The ideal parameters for a drug candidate were defined in Lipinski rule of five (also known as Pfizer’s rule of five) such as molecular weight, logP, number of hydrogen bond donors and number of hydrogen bond acceptors. These characteristics were determined with DruLiTo tool where alkannin (3) and shikonin (4) with lower Logp value than geldanamycin (2) could exhibit better bioavailability compared to geldanamycin (2). Thus, geldanamycin (2) did not comply with required two parameters of molecular weight and hydrogen acceptor of Lipinski’s rule of five [17]. The data of Lipinski’s rule of five for geldanamycin (2), alkannin (3) and shikonin (4) are presented in Table 1.
The three dimensional structure (3D) crystal structures of Hsp90 (PDB ID; 1YES) along with geldanamycin bound Hsp 90 (PDB ID: 1YET) are retrieved from Research Collaborator for Structural Bioinformatics (RCSB) Protein Data Bank (http://www.rcsb.org/). The crystal structure of 1YET is employed for validation of protocol in docking study. The crystal structure of 1YES is employed for docking alkannin (3) and shikonin (4), to get reliable predictions of ligands bindings. The water molecules were removed from crystal structure using Discovery Studio software followed by the addition of Gasteger charges to targets.
The 2D structures of geldanamycin (2) alkannin (3) and shikonin (4) were retrieved from NCBI Pub Chem data base. Energy of these compounds were optimized using MM2 force field methods and converted to #D format, saved as PDB files using Chem Office 2016 (Cambridge Soft Corporation) and further used for docking.
Previously, binding mode of geldanamycin (2) with Hsp90 was identified through X-ray crystallography [18]. Forthwith, binding mode of geldanamycin (2) with Hsp90 was done using protocol that is used for the docking studies.
PyRx software was engaged as virtual screening software. PyRx includes Auto Dock Vina with Lamarckian genetic algorithm as a scoring algorithm. Initially, docking simulation of geldanamycin (2) was done using Hsp90 (1YET) structure for validation of the protocol. Docking simulations of Hsp90 target 1YES with ligands-ATP, geldanamycin (2), alkannin (3) and shikonin (4) were undertaken using Auto Dock Vina [19,20] for predicting the binding site of Hsp90 and ligands. The target conformation (biomacromolecule) was set as rigid unit while the ligands were conceded to be flexible and adoptable to the target. Vina explored for the lowest binding affinity conformations and provided nine different conformations for each 1YES. The lowest binding energy docking poses of each compound were selected. Auto Dock Vina was processed using exhaustiveness of eight and grid box with the dimensions for center of x= 33.934, y= -47.5359, z= 61.6928 with a size of X= 43.0922, Y= 46.3722 and Z= 56.9586 for 1YES. Py Mol v1.3 (Schrodinger, New York, NY, USA) and Chimera (UCSF, San Francisco, CA, USA) were chosen for visual inspection and preparations. The protein-ligand interactions were analysed with the aid of Chimera and Lig Plot v.4.5.3. [21].
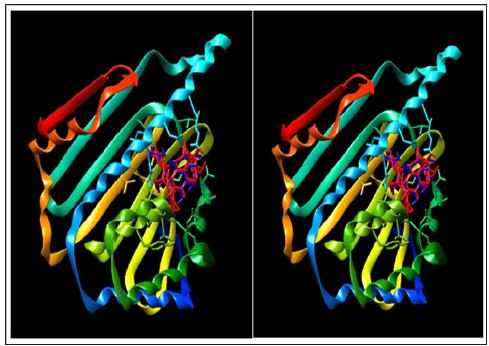
Earlier to docking procedure, the protocol was validated by re-docking extracted ligand, geldanamycin (2), to 1YES and 1YET targets using a previously reported procedure [22]. The target/ligand interactions were monitored by comparing target ligand interactions with 1YET- geldanamycin crystal complex structure with support of Lig Plot. Lig Plot results revealed that the X-ray structure of 1YET complex and protein/ligand docked complexes of 1YET as well as 1YES were shown to be similar, validating the protocol. The protein / ligand interactions of Lig Plot are displayed in Figure 1 and graphical presentation of docking structure in Figure 3. In order to ascertain the putative binding site of alkannin (3) and shikonin (4) in N-terminal domain of Hsp90, NT “blind” docking method was selected using Auto Dock Vina. The entire N-terminal domain was chosen to search for the lowest possible binding energy considering that geldanamycin (2) binds to different location than ATP binding site in cavities of protein [23,24]. Alkannin (3) and shikonin (4) satisfied Lipinski’s rule of five, and exhibited best binding affinity which was comparable to reference ligand, geldanamycin (2) (Figure 4).
Figure 3: Lig Plot showing interactions of Hsp90 (1YET) - geldanamycin (2) X-Ray crystal, docked geldanamycin (2) on Hsp90 (1YET) and docked geldanamycin (2) on Hsp90 (1YES) for validation of protocol.

Figure 4: A: X-Ray structure of Hsp90 (1Yet)-geldanamycin (2) crystal structure and B: X-Ray structure of Hsp90 (1Yet)-geldanamycin (2) crystal structure superimposed on Hsp90 (1YES) docked geldanamycin (2), respectively.
The results from the docking study revealed that ATP (1) conceived interactions through five hydrogen bonds with Asn51, Lys 58, Asp93, Gly137 and Phe138; allosteric interactions through Ala55, Met98, Leu107, Gly132, Gly135, Val136, thr184 and Val186 to the N-terminal domain of Hsp90, with binding energy of -8.4. Geldanamycin (2) bridged two hydrogen bonds with Lys204 and Leu220; allosteric interactions through Leu 89, Val, 207, Lys208, Glu212, Ile218 and Thr219 to N-terminal domain of Hsp90 with binding energy of -6.3. Alkannin (3) bridged five hydrogen bonds with Ser52, Asp54, Asp93, Gly97 and Thr184; allosteric interactions through Asn51, Lys58, Ile96, Thr152, Val186, Met98, Ala55, Leu107, Phe138 and Val150 to N-terminal domain of Hsp90 with binding energy of -7.1. Shikonin (4) bridged two hydrogen bonds Thr184 and Asn106; allosteric interactions Asn51, Ala55, Ile,96, Gly,97, Met98, Leu107, Lys112, Gly135, Val136, Gly137, Phe138 and Tyr139 to N-terminal domain of Hsp90 with binding energy of -7.2.
`Figure 5: Lig Plot-Interactions of ATP (1), geldanamycin (2), alkannin (3) and shikonin (4) with Hsp90 active sites.
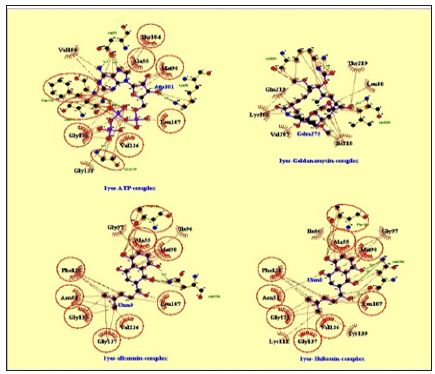
Protein / ligand interactions of Lig Plot are displayed in Figure 5 and graphical presentation of docking structure of ligands, geldanamycin (2) alkannin (3) and shikonin (4) on Hsp 90 protein is displayed in Figure 5. This docking study suggested that both alkannin (3) and shikonin (4) intercalate with N-terminal of Hsp90 with lower binding energies compared to geldanamycin (2), in blind docking studies. Furthermore, shikonin formed two hydrogen bonds whereas alkannin (3) formed five hydrogen bonds with Hsp90 suggesting that alkannin (3) could perform better as Hsp90 inhibitor (Figure 6). The current docking study results hypothesize potentials of alkannin (3) and shikonin (4) as inhibitors of Hsp 90 and further in vitro and in vivo laboratory studies are needed to substantiate these findings.
Heat shock proteins (HSPs) stabilise a number of proteins required for tumor growth. Heat shock protein 90 (Hsp90) is one of the most common of heat-related proteins. Interestingly, a large number of Hsp90-client proteins play significant roles in determining the cancer cells characteristics. Hsp90 and its co-chaperones are essential for the function of these tumor-promoting client proteins. The mechanism-based use of Hsp90 inhibitors alone or in combination with other anticancer drugs should amplify the treatment of various cancers. Hence, inhibition of Hsp90 by particular inhibitors such as geldanamycin (2) and its derivatives decreases the tumor progression. The molecular docking studies with geldanamycin (2), alkannin (3) and shikonin (3) into the binding cavity of Hsp90 protein suggested that shikonin has more binding affinity than geldanamycin (2) with better binding energy. The results of present study can be useful for designing and developing new Hsp 90 inhibitors, beneficial to cure/manage various cancers in future. Further studies using in vitro and in vivo experiments substantiate these findings is required.


