Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Ghazaleh Sadeghi1, Shahin Najar Pirayeh1, Mojgan Forutan2 and Amin Talebi Bezmin Abadi*1
Received: February 01, 2018; Published: February 15, 2018
Corresponding author: Amin Talebi Bezmin Abadi, Department of Bacteriology, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
DOI: 10.26717/BJSTR.2018.02.000763
Background and aims: Helicobacter pylori (H. pylori) is a colonizer in the gastric mucosa of more than 50% of the human population. Complications due to this bacterium include chronic gastritis, gastric mucosa-associated lymphoid tissue lymphoma, peptic ulcer and gastric adenocarcinoma. The aim of this study was molecular detection of H. pylori in biopsy samples by amplification of conserved sequences of dupA gene.
Materials and Methods: The in silico study for detection of conserved region in dupA was performed. Seventy-nine biopsy samples were collected and DNA was extracted from them. The polymerase chain reaction (PCR) was performed to amplify dupA conserved region. The glmM gene was also amplified as a standard control alongside the dupA gene.
Results: Results of 79 samples, 31 and 48 were collected from males and female patients, respectively. The age range of patients was 13-85 years; Mean±SD (49.68±18.82).The patients' complications included 35.4% reflux (13.92% males and 21.52% females), 32.9% gastritis (7.59 males and 25.31% females) and 8.9% peptic ulcer (2.58 males and 6.32% females). The PCR amplification of dupA showed 98% and 100% sensitivity and specificity, respectively, whereas results for glmM were 26% and 100%, respectively.
Conclusion: The findings exhibited that the amplification of dupA conserved region with a high sensitivity and specificity can be considered as a rapid diagnostic method for H. pylroi identification in biopsy specimens.
Keywords: Helicobacter pylori; biopsy samples; Diagnosis; dupA gene; glmM gene
Abbreviations: RUT: Rapid Urease Test; UBT: Urea Breath Test; SAT: Stool Antigen Test
Helicobacter pylori (H. pylori) colonizes half of the human population worldwide and is known as the causative agent of gastric ulcers and adenocarcinoma, chronic gastritis, gastric mucosa-associated lymphoid tissue lymphoma [1]. Diagnosis of H. pylori is performed by several methods each with own advantages, disadvantages and limitations. A classic route to categorize the methods is dependent upon whether or not an endoscopy is essential. Those tests based on biopsy include culture, histological evaluation, PCR and the rapid urease test (RUT), all of which are conducted on tissue obtained during endoscopy. As an alternative way, the urea breath test (UBT), stool antigen test (SAT) and serology can be performed as non-invasive procedures. A second way for classification of these tests is according to if they are used before or after H. pylori eradication treatment [2].
In the histological examination, in which both antrum and corpus biopsies may be required, critical information regarding the mucosa (e.g., presence and severity of inflammation, dysplasia, glandular atrophy, intestinal metaplasia and neoplasia) will be provided [3-6]. The sensitivity and specificity of histology vary from 53% to 90%, relating to the pathologist's experience and density of H. pylori colonization. Increase in the number of biopsies and application of specific stains can enhance the sensitivity of this examination [7]. Several other Helicobacter species may be also present in gut and misdiagnosed with H. pylori. Fluorescent in situ hybridization (FISH) is a new method for specific characterization of H. pylori features, but this method is laborious, expensive, and not utilized in clinical practice [8-9]. The culture method is ideal when using a recently obtained gastric biopsy specimen because of low number of flora in most of cases, having sensitivity of 90% and specificity of 100% if fulfilled under optimal conditions [10].
However, for the delicateness of H. pylori and needing to culture as soon as possible, always it is not true. For this purpose,biopsy samples can be taken in a transport medium (e.g., Stuart's medium) for up to 24 h at 4oC. Culture of H. pylori is performed on several media such as Skirrow agar, Pylori-agar, Wang media, and others with specific antibiotics to inhibit bacterial body flora, and nonselective media (e.g., blood agar, Columbia blood agar, and others). Culture is the most specific method, but depends on personal experience, specimen quality, and use of transport media [11]. It is in general not considered a routine diagnostic way and is not available in most of medical institutions worldwide. Sometimes the method is conducted when antibiotic susceptibility testing is required [2].
PCR allows researchers to detect the bacterium even in very low numbers, however there the possibility that dead bacterial population be present in the gut mucosa and give positive result in this test. PCR results are faster than many other diagnostic methods for identification of bacterial genotypes and when epidemiological studies are performed [12-14]. Some other diagnostic methods have been applied for H. pylori such as urease breath test and stool antigen tests may also be performed. The aims of this study were detection of H. pylori prevalence and evaluation of it in biopsy samples by amplification of gut mucosa and give positive result in this test. PCR results are faster than many other diagnostic methods for identification of bacterial genotypes and when epidemiological studies are performed [12-14]. Some other diagnostic methods have been applied for H. pylori such as urease breath test and stool antigen tests may also be performed. The aims of this study were detection of H. pylori prevalence and evaluation of it in biopsy samples by amplification of conserved region of dupA gene.
The in silico study , For this purpose, a pilot study was performed until June 2015. Six-hundred and seventy-four various sequences of dupA gene were entered to the Seaview (Version 4.5.4) software and were assessed initially (puzzling). For detection of conserved region in dupA, 100 preferred sequences with more overlapping regions were adopted (Figure 1). The PCR was designed using specific primers for the overlapping region.
Figure 1: Detection of conserved region in dupA gene.

Biopsy Samples Collection: Seventy-nine biopsy samples were collected from patients with various complications including indigestion, stomach pain, nausea, reflux, inflammatory bowel disease, gastritis, fatty liver disease, duodenal ulcers, gastric ulcers and other symptoms by gastroenterologist physician. The specimens were transported to the laboratory in micro-tubes containing tyoglycolate medium and stored at -80°C until use in the test.
DNA extraction: DNA was extracted from biopsy samples by utilizing YTA Genomic DNA Extraction Mini kit (Cat No: YT9030) according to the manufacturer's instructions. The kit contains TG1 and TG2 buffers, proteinase k, mini-TG column, W1 buffer, collection tube, wash buffer, elution tube and micro pestle (for tissue homogenization).
PCR amplification of dupA and glmM genes: PCR technique was performed to detect dupA conserved region to amplify 110bp size product. The glmM gene was also amplified as a standard control alongside dupA gene for the amplification of a product with 294bp size. The specific primers included:glmMF: 5'AAGCT- TTTAGGGGTGTTAGGGGTTT3’,glmMR:'5AAGCTTACTTTCTAA- CACTAACGC3’, dupAF: 5’CATGGCGTTTCAAAAAATATCTCAA3’, and dupAR: 5'TTCATCAGTATCTTTTGTGGGGTA3'. PCR conditions for dupA gene included as following: 93°C for 60s, 61°C for 60s, and 72°C for 60s. The thermal profile conditions for glmM were similar to those for dupA gene and both were run for 35 cycles. The strain 26695 was the reference strain used in this assay.
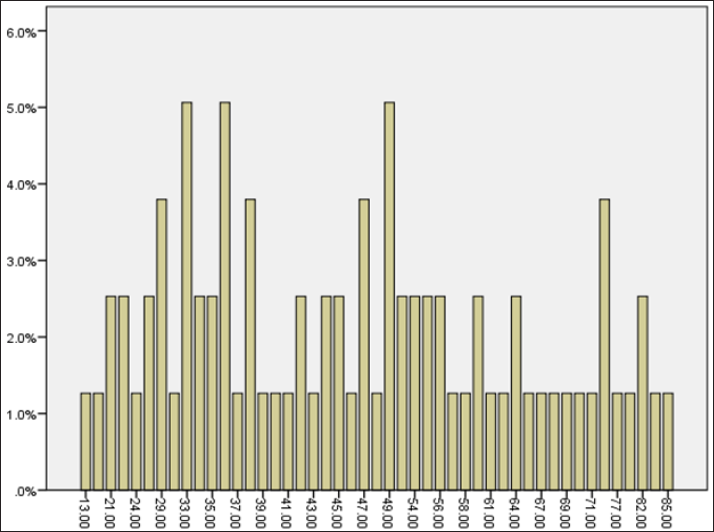
Thirty-one and 48 patients were males (mean age = 47.70±1.37) and females (mean age= 17.39±0.47), respectively. The age range of patients in this study has been demonstrated in (Figure 2). The patients' complications included 35.4% reflux (13.92% males and 21.52% females), 32.9% gastritis (7.59 males and 25.31% females) and 8.9% peptic ulcer (2.58 males and 6.32% females). The ages of 33, 36 and 49 years were predominant age groups. Twenty- three patients had a history of gut infection, but 56 were healthy individuals.
Figure 2: The age distribution of patients in this study.
The PCR amplification of dupA showed 98% and 100% sensitivity and specificity, respectively, whereas results for glmM were 26% and 100%, respectively. The relation between dupA and glmM and gastrointestinal diseases was not significant (p value= 0.443). Therefore, H. pylori was not considered as the only gastrointestinal disease.
The accurate diagnosis of H. pylori is a critical and effective factor for management of many gastrointestinal diseases. Several invasive and non-invasive methods are available in the H. pylori and each has its advantages and drawbacks. These methods have sensitivity and specificity of approximately 90%. Choosing each method is according to the availability and patient's conditions. The invasive methods include endoscopy, culture, rapid urease test and some molecular methods. Urea breath test, fecal antigen test and serology are among those non-invasive methods [15]. PCR method has a higher sensitivity and specificity which can cover many other queries such as needing to rapid and non-invasive diagnosis, determination of mutations related to the antibiotic resistance alongside virulence factors encoding genes, and genotyping [1618].
Several genes have been amplified for the diagnosis of H. pylori such as 16srRNA, ureA, glmM, babA2, rpoB, ssA, cagA, hopQ, icd and PPIase [19-20]. The ureC gene also called glmM encodes the phosphor-glucoseamin mutase, is widely used for the H.pylori diagnosis [21]. We determined sensitivity and specificity of 98% and 100% for dupA and 26% and 100% for glmM genes, respectively among biopsy samples. The glmM low sensitivity makes a drawback in the diagnosis of H. pylori [22]. The dupA gene placed in a plasticity zone of H. pylori genome, have shown to be related to duodenal ulcers [23-25].
Two studies assessing different diagnostic methods in specific settings revealed that histology was the most accurate test: Choi et al. [27] evaluated several methods for H. pylori detection in patients with peptic ulcer which 157 were bleeding and 247 were nonbleeding. The sensitivity and specificity of histology were 92.5 and 96%, and 85 and 92% for rapid urease test (RUT), 40 and 100% for culture, and 97 and 56% for serology, respectively. The culture and RUT sensitivities were significantly higher in non-bleeding patients compared to bleeding patients. By contrast, histology was valid and accurate regardless of bleeding. Tian's study [27] demonstrated in a meta-analysis evaluating diagnostic methods of H. pylori in patients with partial gastrectomy.
The sensitivity and specificity included 93 and 85% for histology, 79 and 94% for RUT and 77 and 89% for urea breath test (UBT), respectively. Their study concluded that histology method was the most accurate test in this setting. In this study, female patients and age groups of 33, 36 and 49 years were more infected with H. pylori which was somewhat in accordance with Gimire's study in which most of patients infected with H. pylori were in 21- 40 years old, but their study showed no association with gender (55.8% male versus 44.2% female) of patients [28]. Kadhim reported a significant difference between male and female patients infected with H. pylori [29]. In our study, the patients' complications included 35.4% reflux (13.92% males and 21.52% females), 32.9% gastritis (7.59 males and 25.31% females) and 8.9% peptic ulcer (2.58 males and 6.32% females).
Nordenstdt et al showed that among 491 individuals, 40.7% had gastritis and 20.5% (n=41) were H. pylori negative [30]. In a study by Abrams, the incidence of reflux-related cardia cancer showed gradually increased, whereas cardia cancer caused by H. pylori has decreased progressively since the mid-20th century [31]. In a study by Franklin, 44.9% (35/78) of adolescent patients with H. pylori exhibited chronic gastritis [32]. In this investigation, no significant relation was observed between the presence of dupA and glmM genes and clinical findings including gastritis, peptic ulcer and reflux. This is in agreement with studies from Brazil and South Africa [33,34].


