Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Samuel Sorie1, Isatta Wurie1*, Umu Khuntum Tejan Jalloh2, Babatunde M Duduyemi1 and Mohamed Samai1
Received: January 29, 2025; Published: February 07, 2025
*Corresponding author: Isatta Wurie, College of Medicine and Allied Health Sciences, University of Sierra Leone, Freetown Sierra Leone
DOI: 10.26717/BJSTR.2025.60.009462
Background: Gastrointestinal infections are a major cause of public health issues, morbidity, mortality and economic
loss. Diarrheal disease disproportionately affects developing nations but also remains a significant concern
in industrialized countries. The aetiologic agents include a wide range of bacteria, viruses, and parasites.
Given the limited research on the prevalence of enteric pathogens in Sierra Leone, this study aimed to evaluate
the prevalence of these pathogens at Connaught Hospital, the main referral tertiary hospital in the country. The
study employed the FilmArray gastrointestinal panel and validated the results against established conventional
PCR methods. Specifically, the study assessed the performance of the gastrointestinal (GI) panel for the simultaneous
detection of six targets: Shigella spp./enteroinvasive Escherichia coli (EIEC), Shiga toxin-producing Escherichia
coli (STEC) (stx1 & stx2), Salmonella spp., Norovirus, and Clostridioides difficile (C. difficile) from stool
specimens.
Objective: To compare the performance of FilmArray GI panel in detecting Shigella spp./EIEC, STEC (stx1 & stx2),
Salmonella spp., Norovirus, and C. difficile with the performance of established PCR methods.
Method: Retrospectively collected stool specimens (n=134) and prospectively collected stool specimens (n=38)
from patients with gastrointestinal infection in an existing enteric prevalence study were analyzed using real
time PCR and nucleic acid test (NAT), and GeneXpert as comparison standards, along with the multiplex FilmArray
GI panel. The sensitivity, specificity, positive predictive values (PPV) and negative predictive (NPV) values
were calculated to determine the overall agreement between the multiplex FilmArray GI panel results and these
comparison standards.
Result: The FilmArray GI Panel sensitivity was 100% for both STEC (stx1 & stx2) and C. difficile, and 90%,
88.89% and 77.78% for Norovirus, Shigella spp./EIEC (ipaH) and Salmonella spp. respectively. The FilmArray GI
Panel specificity was >95.1% for both STEC stx1 & stx2 and Salmonella spp. targets, and 92.31%, 88. 24 % and
87.50% for Shigella spp./EIEC (ipaH), C. difficile and Norovirus, respectively.
Keywords: Filmarray; Quality Assurance; Sensitivity; Specificity
Abbreviations: C: Comparison standard; DNA: Deoxyribonucleic acid; EAEC: Enteroaggregative E. coli; EIEC: Enteroinvasive E. coli; E. coli: Escherichia Coli; FA: FilmArray; FN: False Negative; FP: False Positive; GI: Gastrointestinal; IPC: Internal Positive Control; NPV: Negative Predictive value; PCC: Probe Check Control; PCR: Polymerase Chain Reaction; PPV: Positive Predictive Value; QC: Quality Control; RNA: Ribonucleic Acid; C. difficile: Clostridium difficile; IpaH: Invasion Plasmid Antigen H Gene Sequence; Salmonella spp: Salmonella species; SPC: Sample Processing Control; STEC: Shiga – Toxin Producing Escherichia coli; stx1: Shiga Toxin 1; stx2: Shiga Toxin 2; TN: True Negative; TP: True Positive; Shigella spp./EIEC – Shigella species/ Enteroinvasive Escherichia coli
Infectious gastroenteritis is a leading cause of morbidity and mortality worldwide, especially in Africa. (Liu, et al. [1]) This is largely due to inadequate water treatment, insufficient food safety measures, and poor sanitary conditions (Liu, et al. [1]). Acute diarrheal disease poses a significant health risk that can be life-threatening and capable of causing outbreaks. Hence, swift laboratory confirmation and timely initiation of the appropriate therapy for these diseases are crucial. The main aetiologic agents of infectious gastroenteritis include bacteria, viruses, and parasites. Transmission of enteric pathogens occurs mainly through faecal-oral pathways (Wagner, et al. [2]). Ingestion of contaminated food and water, as well as contact with faecal contaminated environments, have been identified as the key transmission pathways for bacterial and protozoan enteric pathogens (Berendes, et al. [3]). Although enteric viruses can be transmitted through similar routes, person-to-person transmission is also a significant transmission pathway.
In many developing nations, disease diagnosis primarily relies on symptoms (Glass, et al. [4]). For example, acute persistent rice water stool is associated with cholera, while bloody diarrhoea is linked with dysentery (Glass, et al. [4]). However, relying solely on clinical symptoms for a definitive diagnosis is inadequate, as diarrhoea is a common manifestation of infectious gastroenteritis, regardless of aetiology (Thielman, et al. [5]). Identifying the specific causative agent is essential for disease surveillance, prevention and control. Therefore, effective tools and procedures for diagnosis are essential to optimize treatment and prevention interventions. Given the variety of pathogens that cause gastroenteritis, most laboratories use multiplex polymerase chain reaction (PCR) to test for pathogens in stool (Keusch, et al. [6].
A 2014 US study, “Multicenter Evaluation of the BioFire FilmArray Gastrointestinal Panel for Etiologic Diagnosis of Infectious Gastroenteritis,” compared the FilmArray GI Panel’s results from stool samples to those obtained using standard stool culture and molecular methods. The FilmArray GI Panel correctly identified 100% of positive cases for 12 out of the 22 targets, and over 94.5% for 7 more of the 22 targets. The test also showed high accuracy in identifying negative cases, with a specificity greater than 97.1% for all targets on the panel (Pawlowski, et al. [7]). An ongoing study at Connaught Hospital in Freetown, Sierra Leone, is utilizing a multiplexed molecular assay, the FilmArray Gastrointestinal (GI) Panel to assess the prevalence of enteric pathogens at the facility. The results obtained from the FilmArray, however, need to be evaluated against the quality assured assays as comparison standards to ensure their accuracy and reliability. In this study, the accuracy of results from the multiplex FilmArray GI panel will be compared to real-time PCR for three common bacterial pathogens (Shiga toxin-producing Escherichia coli (STEC), Salmonella spp., and Shigella spp./enteroinvasive Escherichia coli (EIEC)) and to the GeneXpert platform for Norovirus and Clostridium difficile. These specific pathogens were chosen for quality assurance because they pose a great public health threat, and quality assured assays are readily available.
This research project was part of a broader prevalence study conducted at the Connaught Hospital Laboratory, specifically within the bacteriology and molecular units. The study aimed to determine the prevalence of enteric pathogens among patients with diarrhoea admitted at the hospital. In this research project, the design was such that to provide quality assurance of the results produced by the FilmArray GI panel by comparing the results through diagnosis of the same stool specimen used in the enteric prevalence study but now using those results as the comparison standards. This study was conducted from April to July 2019 and included 172 admitted adult patients with diarrhoea stool samples. Recruitment of participants was carried out by nurses trained in Good Clinical Practice (GCP).
Specimen
Patients were instructed to collect their stool specimens in a 50ml clean, wide-necked, leakproof stool collection container. Specimens that met the following inclusion criteria were selected for the study:
• The specimen matched the description of diarrhoea faeces, including
watery, bloody or rice-water stools.
• The laboratory received the specimen with a request form for
stool culture.
• The specimen was sufficient for testing using the FilmArray GI
Panel, PCR and GeneXpert.
For ethical considerations, each sample a unique identification number (SCN) separate from any personal details. This number was linked to general information about the patient, such as age, sex, whether they were in the hospital (inpatient, outpatient, or ER), and the date the sample was taken. All testing and analysis used only the SCN-labeled samples, so no one working with the samples could see any information that could identify the patient.
Upon receiving the stool specimens, laboratory personnel immediately cultured the specimens on the appropriate media as part of the routine clinical procedure, monitoring the culture until a diagnosis was made. 2ml of each specimen was transferred into sterile containers for FilmArray testing. Two additional aliquots of the stool specimen were made in 2ml cryovial tubes- one containing a neat sample and the other preserved with RNA later solution, which preserves the nucleic acid for PCR diagnosis and GeneXpert testing. The cryovial tubes were labelled with the sample’s unique ID number, date, lab personnel’s initial and “RNA” on the tubes containing RNA later. The remaining stool sample was stored in the original stool collection tube in a refrigerator at 2-8°C, while the two aliquots were stored in a -80°C freezer until testing by PCR and GeneXpert was conducted. The results of other tests ordered by the clinicians were not collected or utilized in this study. The majority of the stool specimens (77.9%) analyzed were archived samples collected and stored at -80oC freezer during the first phase of the enteric prevalence study, and 22.1% were prospectively collected and analyzed.
Shiga toxin-producing E. coli, EIEC, and Shigella spp
The important virulence factors of STEC are Shiga toxins 1 and 2, which are encoded by the stx1 and stx2 genes. These genes are located in lambdoid bacteriophages integrated into the bacterial host genome (Buss, et al. [8]). In this study, the PCR assay Altona RealStar EHEC PCR Kit 2.0 (Altona Diagnostics [9]) a qualitative in vitro diagnostic test was used for the detection and differentiation of DNA specific for both stx1, and stx2 of Escherichia coli, the invasion plasmid antigen H (ipaH) of enteroinvasive Escherichia coli (EIEC), and Shigella spp.
Salmonella spp: The Microseq (Applied Biosystems, Thermofisher, Massachusetts, USA) PCR method was used for the detection of specific targets for Salmonella spp.
Norovirus: Norovirus has seven known genogroups. However, majority of Norovirus infection in humans are caused by genogroup I and II viruses. The GeneXpert Norovirus assay recently received FDA clearance for the detection and differentiation of Norovirus genogroups I and II (GI and GII), which account for the vast majority of infections in humans (Friedrich, et al. [10]). This study employed the GeneXpert Norovirus assay for the detection and differentiation of Norovirus genogroups I and II (GI and GII) as a comparison standard.
Clostridium difficile: The GeneXpert C. difficile assay (Xpert CD assay; Cepheid, Sunnyvale, CA, USA) was used as the comparison standard in this study.
The above methods were used as comparison standards to the FilmArray. This aims to provide quality assurance of the FilmArray results in order to determine if the FilmArray results are in concordance with established molecular PCR diagnostic tests. Given that the FilmArray is a relatively new device, with limited validation data, it is crucial to assess its accuracy.
Film Array in Perspective
The FilmArray Gastrointestinal Panel (GI) is a multiplex in-vitro diagnostic test used for the simultaneous and rapid (1 hr) detection of 22 pathogen targets directly from stool samples known to cause gastrointestinal infections. Out of the 22 pathogen targets detectable by FilmArray GI panel, 5 were selected for quality assurance in this study. These include, Salmonella spp., Shiga toxin producing E. coli (STEC), Shigella spp., Norovirus., and Clostridium difficile. The performance of FilmArray to detect these targets was assessed.
Clostridioides difficile (formerly known as Clostridium difficile) toxin A/B
The Film Array GI Panel uses one comprehensive test to find Clostridioides difficile strains that produce toxins. It looks for both the gene that makes toxin A (tcdA) and the gene that makes toxin B (tcdB). While most toxin-producing strains make both toxins, finding either gene means the strain can cause disease. Empirical testing and in silico sequence can find all known toxin types. However, FilmArray can’t tell the difference between toxin A and toxin B. If it finds either or both toxin genes, the result is reported as “Clostridioides difficile toxin A/B Detected.
Diarrheagenic E. coli/Shigella pathotypes
The FilmArray GI Panel is able to identify the genetic markers of common E. coli and Shigella strains that cause diarrhea. If the test shows multiple types of these bacteria, it could mean there are several different strains present, or just one strain that has the genetic features of more than one type. For example, the 2011 E. coli O104:H4 outbreak strain had characteristics of both Shiga toxin-producing E. coli (STEC) and Enteroaggregative E. coli (EAEC).
shiga-like Toxin-Producing e. coli (stec) Shiga-Like Toxin Genes 1 and 2 (stx1/stx2)
The FilmArray GI Panel uses two tests (STEC 1 and STEC 2) to find the genetic material for Shiga-like toxin 1 (stx1) and Shiga-like toxin 2 (stx2). The test results don’t specify which toxin(s) were found. If either or both tests are positive, the result will be reported as “Shiga- like toxin-producing E. coli (STEC) stx1/stx2 Detected.” Because Shiga toxin (stx, which is the same as stx1 in STEC) is also present in Shigella dysenteriae, a sample that tests positive for both STEC stx1/ stx2 and Shigella spp./Enteroinvasive E. coli (EIEC) could mean that S. dysenteriae is present.
E. Coli o157:
To help find STEC of the O157 type, the FilmArray GI Panel has a specific test (EC O157) that looks for a gene only this type has. While some E. coli O157 strains don’t have the Shiga-like toxin genes, and it’s unclear how harmful these non-STEC strains are, the O157 test result is only reported if a Shiga-like toxin gene is also found. If both STEC stx1/stx2 and E. coli O157 are detected, the O157 result is reported as “Detected.” If STEC stx1/stx2 is not detected, the O157 result is marked as N/A (Not Applicable). The FilmArray GI Panel cannot tell the difference between an infection with just one toxin-producing STEC O157 and a rare case where a STEC (not O157) with toxins is present along with an E. coli O157 that doesn’t have the toxin genes.
Shigella/Enteroinvasive E. coli (EIEC):
The FilmArray GI Panel contains a single assay for the detection of ipaH, a gene unique to all Shigella species, as well as Enteroinvasive E. coli (EIEC). This method cannot differentiate between Shigella and EIEC, so detection of ipaH will result in a “Shigella/Enteroinvasive E. coli (EIEC) Detected” test result. Shiga toxin (stx) is found in Shigella dysenteriae; therefore, positive test results for STEC stx1/stx2 and Shigella/Enteroinvasive E. coli (EIEC) in the same sample may indicate the presence of S. dysenteriae.
Norovirus Gi/Gii
The FilmArray GI Panel uses two tests (Noro 1 and Noro 2) to find the most common types of norovirus that infect humans (GI and GII). It won’t detect genogroup GIV, noroviruses that infect animals, or similar viruses like Sapovirus. The results don’t specify whether GI, GII, or both were found. If either or both tests are positive, the result is reeported as “Norovirus GI/GII Detected.
Each FilmArray GI Panel kit contained individually packaged Bio- Fire GI Panel pouches, single-use sample buffer ampoules, single-use pre-filled hydration injection vials, single-use sample injection vials, and individually packaged transfer pipettes. In this study, the sample preparation and loading process involved several key steps to ensure accuracy and reliability. Initially, the pouch was securely placed into the Pouch Loading Station, with the Sample Injection Vial positioned in the red well and the Hydration Injection Vial in the blue well. The hydration of the pouch was achieved by twisting off the cap of the Hydration Injection Vial and inserting it into the pouch hydration port. The vial was then forcefully pushed down to puncture the seal, allowing the hydration solution to be drawn into the pouch. For the sample mix preparation, the Sample Buffer was added to the Sample Injection Vial by inverting the Sample Buffer Ampoule, pinching the plastic tab to break the seal, and dispensing the buffer into the vial with a controlled squeeze. The stool specimen, thoroughly mixed in its transport media, was then added to the Sample Injection Vial using a transfer pipette. The vial was securely closed and gently inverted three times to ensure proper mixing. The Sample Injection Vial was returned to the red well of the Pouch Loading Station. Finally, the sample mix was loaded into the pouch by unscrewing the Sample Injection Vial from its cap, pausing briefly, and then inserting the vial into the pouch sample port. The vial was pushed down to puncture the seal, allowing the sample mix to be drawn into the pouch. These steps were meticulously followed to prepare and load the samples for subsequent analysis.
Inside the FilmArray pouch, the genetic material (nucleic acids) is released from the sample using a combination of physical and chemical breakdown. Then, standard magnetic bead technology is used to purify the genetic material. Once the genetic material is extracted and purified, the FilmArray uses a two-stage nested multiplex PCR process to amplify specific DNA sequences. In the first stage, reverse transcription and first stage PCR occur in blisters ‘‘F’’ and ‘‘G’’ (Figure 1). The PCR master mix, which contains the reverse transcriptase, is transferred from well 7 of the fitment into blister ‘‘F’’. A mechanical hot start is achieved by holding the contents of the two blisters separate with a hard seal until they reach 54oC. Reverse transcription occurs during the initial 3-minute hold at 54oC. The first stage PCR consists of 26 cycles of heating to 94oC for 4 seconds, followed by 60oC for 19 seconds. At the end of the cycling process, the reaction is diluted approximately 225-fold into the second stage PCR master mix by two successive dilution steps, first with TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0) from well 8 and then with PCR master mix from wells 9 and 10, and a fluorescent double-stranded DNA binding dye (LC Green® Plus, BioFire). The mixture is then spread across the wells of the array. Each well contains specific primers designed to amplify DNA sequences within the PCR products created in the first stage of the PCR reaction.
The second stage PCR, or nested PCR, is performed in a singleplex fashion in each well of the array. The conditions for this stage involve 30 cycles of heating to 94 °C for 4 seconds followed by cooling to 63 °C for 19 seconds, with ramp rates of approximately 1.7 °C/sec. After the second stage PCR, a melt curve analysis is performed to detect signature amplicons that indicate the presence of specific targets. The sample is held at 63 oC for 5 seconds, followed by a linear ramp in temperature from 68 oC to 95oC at a nominal rate of 0.5 °C/ sec. A digital camera captures fluorescent images of the PCR reactions, and software interprets the data. The FilmArray software automatically analyzes the DNA melting curves from each test and combines these results with the internal quality control results from the pouch to give a final test result for each organism included in the panel. This analysis begins with the control assays. If the controls yield a positive result, the analysis proceeds to the pathogen assays, and the results are reported. If control assays return a negative result, the run is declared ‘Invalid’, and no organism results are reported.
Real-Time PCR Comparator Analysis
The PCR procedure was performed at the Molecular Unit of the Connaught Hospital laboratory. Stool samples were inactivated, and DNA was extracted before amplification using the Lightcycler 96 thermocycler.
Sample Lysis
Lysis of stool samples was performed in a biosafety cabinet, following the appropriate biohazard guidelines for working with infectious pathogens. Lysis buffer (AL buffer) from the Qiagen QIAamp Fast stool DNA mini kit was used to lyse samples following a pre-treatment with InhibitEX buffer, which lyses human cells in the stool specimen and separates DNA-degrading substances and PCR inhibitors present in the stool sample from the DNA. This sample matrix was then pelleted by centrifugation, and the DNA in the supernatant was added to Proteinase K, which digests and degrades proteins during a 700C incubation. AL buffer was subsequently added to the lysed bacteria cells.
Deoxyribonucleic Acid Extraction
Bacterial DNA was manually extracted from the stool specimen using the QIAamp Fast DNA Stool Mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. DNA extraction was done in batches of 12 and above (see annex for DNA extraction SOP). For the comparator analysis, extracted DNA was analyzed using specific PCR assays for the target pathogens. The PCR assay; ‘EHEC PCR Kit 2.0’ by Altona Diagnostics, a qualitative in vitro diagnostic assay was used for the detection and differentiation of DNA specific for Shiga toxin 1 (stx1), and Shiga toxin 2 (stx2) of Escherichia coli and the invasion plasmid antigen H (ipaH) of EIEC, and Shigella spp. The assay included a negative control, positive control, and a heterologous amplification system (internal control) to identify potential PCR inhibition and confirm the integrity of the reagents in the kit. The probes specific to stx1 DNA are labelled with a fluorophore similar to Cy5, those specific to stx2 DNA were labelled with a fluorophore FAM, and those specific to ipaH DNA are labelled with the fluorophore ROX. The probe specific for the internal control (IC) were labelled with the fluorophore JOE. The kit contained two MasterMix reagents (Master A and Master B), which included all components (PCR buffer, DNA polymerase, magnesium salt, primers and probes) that allowed PCR-mediated amplification and target detection of stx1 specific DNA, stx2 specific DNA, ipaH specific DNA and IC in one reaction setup. A DNA template of 9μl was added to a 21μl mixture of RT-PCR Mastermix reaction (Master A – 5μl, Master B – 15μl, IC -1μl) to give a total reaction volume of 30μl. The cycling conditions included an initial hold stage of 2 minutes at 950C followed by a two-step cycling stage of 15 seconds at 950C and 45 seconds at 580C on the Lightcycler 96 RT-PCR instrument by Roche. A total of 45 cycles were run. A unidirectional workflow technique was used to prevent contamination and ensure the reliability of all laboratory testing. The instrument was used in the mode of a standard run with no passive reference dye, and analysis was conducted with a manual threshold setting.
The Microseq Salmonella spp. Detection PCR kit by Applied Biosystems was used for the detection of specific targets for Salmonella spp. The kit contained lyophilized preformatted assay beads, which held the active enzyme, the target-specific primer and probe set, Internal Positive Control (IPC), and other PCR reagents. The IPC helps prevent false negatives by detecting materials that could inhibit target amplification. The probe for Salmonella DNA detection was labelled with the fluorophore FAM, and the probe for the IPC was labelled with the fluorophore JOE. To hydrate the lyophilized assay beads, 20μl of nuclease-free water was added, followed by 10μl of DNA template to obtain a final reaction volume of 30μl. Serial dilution of known Salmonella spp was performed to assess the efficiency of the PCR assay. DNA extracted from a positive Salmonella spp. Isolate was used as the positive control in each run. Samples showing exponential amplification and a cycle threshold (Ct) value of 35 or less were considered positive.
Efficiency of RT-PCR Assay
A serial dilution of extracted nucleic acid with known cycle thresholds was performed to determine the detection range, maximum analytical sensitivity, and linearity. The cycle threshold (Ct) values obtained from the serial dilution were plotted on a logarithmic scale, along with corresponding concentrations, to generate a linear regression curve. The percentage efficiency of the PCR reaction was then calculated from the slope of the curve. Linearity was measured using the correlation coefficient analysis (R2) for each primer/ probe set master mix. An analysis of the negative and positive control was done to determine the background fluorescence noise and set the threshold used for the analysis of both RT-PCR assays. This was done for all the specific targets.
GeneXpert
The GeneXpert was used as the comparison standard for the diagnosis of Norovirus and C. difficile. The Xpert Norovirus and Xpert C. difficile assays were performed according to the manufacturer’s instructions. Exactly 150μl of the stool specimen was added to the lysis buffer, incubated for 20 minutes, and 1 ml was loaded onto the cartridges for processing on the GeneXpert instrument. Xpert Norovirus and Xpert C. difficile assay cartridges contain a sample processing control (SPC) and a probe check control (PCC). The SPC controls for adequate specimen processing and the presence of PCR inhibitors, while the PCC controls for reagent rehydration, PCR tube filling within the cartridge, probe integrity, and dye stability. All controls had to perform as expected in all cases for the assay result to be considered valid. The PCR results will be considered valid when the positive, negative, and specimen inhibition controls performed as expected.
A FilmArray GI Panel result will be accepted true positive (TP) or true negative (TN) only when it agrees with the result from the comparator method (PCR & GeneXpert). Discordant results would mean false positive (FP) or false negative (FN)
Calculations and Statistical Analysis
Results shows the sensitivity, specificity, positive predictive value, and negative predictive value.
Sensitivity = [no. of TP/ (no. of TP/no. of FN)] *100
Specificity = [no. of TN/ (no. of TN/no. of FP)] *100
PPV = [no. of TP/ (no. of TP/no. of FP)] *100
NPV = [no. of TN/ (no. of TN/ no. of FN)] *100
The study period was between April to July 2019 and 172 stool samples were tested. All the samples (100%) were adequate for testing using PCR and GeneXpert comparison standards. The comparison standards used for the quality assurance of the 172 samples analyzed are shown in Table 1.
Table 2 shows how often each pathogen was detected by the FilmArray GI Panel during the study. Out of 172 samples tested, 91 had at least one pathogen detected, which is a 52.9% positivity rate. Table 3 displays how well the FilmArray GI Panel performed for each individual pathogen it tests for. Sensitivity and specificity were calculated by comparing the FilmArray results to the standard methods listed in Table 1, without investigating any differences in results.
Table 3: Performance summary and characteristics of the FilmArray GI Panel versus comparator assays.
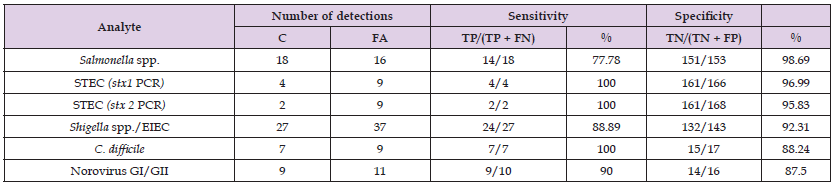
Note: C, comparison standard as defined in Table 1; FA, FilmArray GI Panel.
Most laboratories do not routinely test for pathogens such as Norovirus or C. difficile despite Norovirus having a relatively high prevalence in certain populations (Cardemil, et al. [11]) In this study, both pathogens were detected in 7% of the specimens. Similarly, most laboratories do not currently test for the diarrheagenic E. coli, which is endemic in developing nations and is known to cause endemic bloody diarrhoea (Cardemil, et al. [11]). The FilmArray GI Panel detected EIEC and STEC in 21.51% and 5.20% of all study specimens. This highlights gaps in routine laboratory testing, which can lead to the underdiagnosis of significant gastrointestinal pathogens like Norovirus, C. difficile, and diarrheagenic E. coli strains (EIEC, STEC). This also shows the FilmArray GI Panel can enhance diagnostic capabilities.
There are several limitations of the study that are worth highlighting. The targeted pathogens in the study have limited number of positives which limits the accuracy of the resulting diagnostic accuracy figures but they are a preliminary examination as opposed to a full evaluation of the FilmArray GI panel. The targeted pathogens were few and the number of samples was low (n=172). These may not be representative of the entire country; hence the statistics should be construed with caution. Sample size from previous study on the evaluation of the performance of FilmArray GI panel are high; n=1,500 (Pawlowski, Warren and Guerrant, 2009). Additionally, some gastrointestinal pathogens can be carried asymptomatically, complicating the interpretation of positive results. Detection of organisms by molecular diagnostics may therefore not mean acute infection or the pathogen is responsible for the symptoms the patient is having. For example, both Norovirus (Kirking, et al. [12]) and Salmonella spp (Gonzalez, et al. [13]) can be shed for weeks to months after symptoms subside (Atmar et al. [14]). There is also a possibility of false-negative results due to the presence of sequence variants in the gene targets of the assay, [15] procedural errors, and/or inadequate numbers of organisms for amplification. Additionally, it is possible that the results that were positive by FilmArray but negative by the comparison standards were because of reasons related to the handling and storage of specimens before PCR testing. For example, delays in storing samples at -80°C could have degraded nucleic acids. It is also noteworthy that the FilmArray has multiple targets, and the time and resources required to validate multiple PCR targets are very high and beyond the scope of this study.
In conclusion, this is a preliminary quality assurance study of FilmArray GI panel; and can be used for generating valuable insights concerning the variety of prevalent enteric pathogens circulating in Sierra Leone. The results in this study call for a more comprehensive comparative study to fully evaluate the performance of the FilmArray GI panel. The figures provide useful data that can be used in a larger study.
We would like to thank the College of Medicine and Allied Health Sciences, the University of Sierra Leone, the United Kingdom Public Health Rapid Support Team, the Sierra Leone Ministry of Health and Sanitation’s medical laboratory team and the management and staff of Connaught Hospital Laboratory.
None.


