Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Karine Grigoletto Rossi Cordeiro#, Letícia da Silva Bezerra#, Davi Zanoni Valente, Regiane Priscila Ratti and Larissa Teodoro Rabi*
Received: October 11, 2024; Published:November 08, 2024
*Corresponding author: Larissa Teodoro Rabi, Department of Biomedicine, Nossa Senhora do Patrocínio University Center (CEUNSP), Itu/SP, Brazil
DOI: 10.26717/BJSTR.2024.59.009297
Objective: This study aimed to evaluate possible morpho functional, stability or flexibility alterations resulting from the presence of polymorphisms (SNPs) in the NEDD9 gene and its possible use as biomarkers for acute myeloid leukemia (AML).
Materials and Methods: We used 16 in silico tools to evaluate the impacts on DNA and protein structure, function, flexibility, and stability.
Results: Our analysis showed 12 polymorphisms that promote amino acid changes in protein structure. The rs371007711 (G/A; S512F) was considered deleterious in 5 (83.33%) of the tools evaluated by PredictSNP2.0, indicating changes in the structure and function of DNA, as well as its amino acid alteration, was considered deleterious in all evaluated tools, demonstrating possible changes in the protein structure and function. In addition, a decrease in protein stability was observed. Similarly, rs185882325 (T/C; Y261C) and rs368480197 (G/A; R449C) were considered deleterious in 4 (66.67%) of PredictSNP2.0 tools, suggesting changes in DNA function and structure. Moreover, the amino acid substitutions (Y261C and R449C) were also considered deleterious in all of the evaluated tools, including a decrease in protein stability.
Conclusion: The rs371007711, rs185882325, and rs368480197 polymorphisms may play an important role in AML and can contribute as possible biomarkers for the early diagnosis and/or prognosis of AML.
Keywords: Biomarkers; Acute Myeloid Leukemia; NEDD9; Polymorphisms
Abbreviations: Chr: Chromosome; ID: Identification of the SNP; Ref: Reference Allele; Alt: Altered Allele; D: Deleterious; N: Neutral; U: Unknown; AA: Amino Acid
Several blood-related diseases have been reported to be correlated with intense genetic diversity, mainly caused by chromosomal abnormalities or the presence of single nucleotide polymorphisms (SNPs) or mutations. These alterations can lead to the activation of proto- oncogenes or the inactivation of tumor suppressor genes [1]. Additionally, it is known that SNPs play an important role in identifying the risk of developing diseases and even serving as biomarkers for the diagnosis or prognosis of several conditions, including genetic, autoimmune, and cancer-related diseases [2,3]. Leukemia is characterized as a malignancy of the stem cell precursors in the bone marrow, which can cause changes in early developmental stage cells, facilitating unstable proliferation [1]. According to the National Cancer Institute of Brazil, it is estimated that for each year of the three-year period 2023-2025, Brazil will have 11,540 new diagnoses of leukemia (5,290 in women and 6,250 in men) [4]. Acute myeloid leukemia (AML) is characterized by the alteration of hematopoietic stem cells, causing the proliferation of myeloid blasts [5]. The mechanisms of activation and onset of the disease have not yet been fully elucidated in the literature; however, there is a notable correlation with genetic and epigenetic factors [5,6]. The diagnosis of AML consists of identifying clinical symptoms such as bleeding, recurrent infections, and disseminated intravascular coagulation. In addition, laboratory tests are carried out, such as a blood count [5], in which the main alterations observed are leukocytosis, anemia, and thrombocytopenia [7], as well as the identification of more than 20% of myeloid blasts [5-7]. These observations correspond to an important stage in the diagnostic process, as they confirm suspicions and lead to complementary tests, such as bone marrow examination [5-8], and molecular and cytogenetic techniques [1], which allow the identification of the subtype specific of AML, assisting in the therapeutic’s choice.
The neural precursor cell expressed, negatively regulated by development-9 gene (NEDD9), also known as HEF1 or CAS-L was discovered in 1996 by Golemis [9] and is part of the CAS family of adhesion anchoring proteins [6]. The NEDD9 protein (encoded by the NEDD9 gene) is responsible for complexes of oncogenic kinases, focal adhesion kinases (FAK), regulation of cell signaling cascades, control of several tumorigenesis and metastatic processes [10], participates mainly in the apoptosis pathways, cell cycle and migratory activity and plays an important role in physiological modifier of various types of cancer [10]. Overexpression of the NEDD9 protein is related to a worse prognosis in several types of cancer [6], such as breast [11,12], lung [13,14], prostate [15], colorectal [16,17], and renal cancer [18], and it is been reported to be correlated with worse prognosis in leukemias, especially AML [6]. Its high expression induces cell migration, promoting focal adhesion, which stimulates signaling related to proliferation and genomic instability, possibly contributing to the aggressive behavior of tumor cells [10]. Biomarkers are used for diagnosis, prognosis, predicting response to treatment, or identifying the risk of developing diseases [4]. This makes it possible to align decisions and improve clinical outcomes [6]. The NEDD9 protein has been considered a potential biomarker in several types of cancer, including AML, mainly because its overexpression is related to a worse prognosis [6]. Thus, this study aimed to assess the possible morpho functional, stability, or flexibility protein alterations resulting from the presence of polymorphisms in the NEDD9 gene.
The unsnaps were retrieved from the National Center for Biotechnology Information (NCBI) dbSNP database and UniProt was used to obtain the FASTA sequence of the NEDD9 protein (ID: Q14511). PredictSNP2 [19] was used to evaluate the nucleotide variations resulting from the presence of polymorphisms in the NEDD9 gene. It has five integrated tools, CADD prioritizes functional, deleterious, and pathogenic variants in various categories. DANN can indicate non- linear relationships. FATHMM-MKL integrates functional annotations, based on nucleotide sequence conservation, predicting variant functional impacts of coding and non-coding sequences. FUNSEQ2 prioritizes and annotates somatic modifications, bringing together various resources for genome and neoplasm studies and GWAVA prioritizes non-coding variations. PredictSNP1 [20] was used to assess the impact of amino acid changes in the NEDD9 protein due to the presence of missense polymorphisms in the NEDD9 gene. This is a consensus of integrated tools that promote the assessment of structural and functional impacts on the protein.
SIFT assesses the impact on protein function. PolyPhen-1 uses an empirical method to predict alterations that may or may not affect protein function and PolyPhen-2 assesses the impact on protein function and structure. MAPP analyzes and quantifies the protein's physicochemical variations. PhD-SNP evaluates protein structures and sequences. SNAP evaluates the functional effects on the secondary protein structure and PANTHER analyzes whether the alterations affect protein function. Also, MuPRO was used to identify protein stability alterations, and DYNAMUT was used to assess the impact of amino acid changes on the interaction dynamics with adjacent amino acids in protein structure and it also allows the visualization of protein flexibility.
The data was collected in March/2023. A total of 72.591 polymorphisms were identified for the NEDD9 gene, of which 699 are missense; however, after removing duplicate polymorphisms and selecting only those that were capable of generating amino acid exchange in the protein structure, 12 polymorphisms were obtained for analysis: rs201437141, rs373441149, rs145744844, rs377171913, rs185882325, rs368480197, rs371007711, rs144452810, rs372980300, rs202065635, rs373145975 and rs1803711. The 12 polymorphisms were evaluated according to DNA alterations and protein alterations analysis. The rs185882325 is an exonic polymorphism that alters thymine by cytosine (T/C) in DNA, its alteration was considered deleterious in 4 (66.67%) tools by PredictSNP2 (Table 1) suggesting structural and functional impacts on DNA. In addition, the GWAVA tool did not recognize this alteration. This polymorphism promotes an amino acid change in the protein structure from tyrosine (Y) to cysteine (C) at position 261 (Y261C). The Y261C alteration was considered deleterious in all the tools evaluated by PredictSNP1 (Table 2), indicating the possible structural and functional alteration of the protein. In addition, the MuPRO analysis showed a decrease in the protein's stability (ΔΔG = -0.5591579). In addition, the Dynamut tool showed an increase in the flexibility of the protein (ΔΔSVib ENCoM: 0.178 kcal.mol-1. K-1), but no changes were observed in the binding profile with adjacent amino acids. Similarly, rs368480197, an exonic polymorphism that changes guanine to an adenine (G/A) in DNA, was considered deleterious in 4 (66.67%) tools evaluated by PredictSNP2 (Table 1), indicating possible functional and structural impacts on DNA.
Note: Abbreviations: Chr: Chromosome; ID: Identification of the SNP; Ref: Reference Allele; Alt: Altered Allele; D: Deleterious; N: Neutral and U: Unknown.
Table 2: Analysis of the morpho functional impacts on the NEDD9 protein caused by the presence of polymorphisms in the NEDD9 gene.
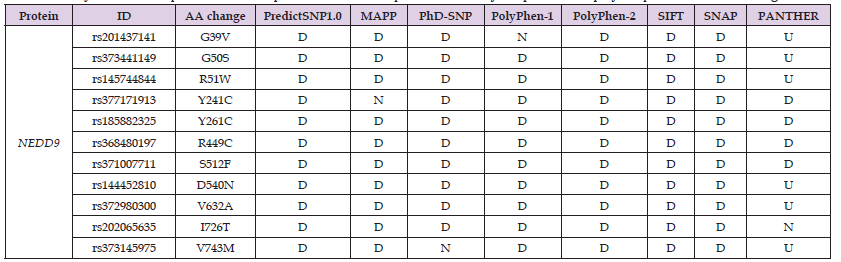
Note: Abbreviations: ID: Identification of the SNP; AA: Amino Acid; N: Neutral and U: Unknown.
However, the GWAVA tool did not recognize this polymorphism. It promotes an amino acid change in the protein structure from an arginine (R) to a cysteine (C) at position 449 (R449C). This change was considered deleterious in all the tools evaluated by PredictSNP1 (Table 2), suggesting a possible alteration in the protein's function and structure. Again, a reduction in protein stability was observed (ΔΔG = -0.26810935; MuPRO). In addition, the Dynamut tool observed an increase in the protein's flexibility (ΔΔSVib ENCoM: 0.059 kcal.mol-1. K-1) and a change in the binding profile with adjacent amino acids due to the appearance of halogen bonds (Figure 1A). The rs371007711 is an exonic polymorphism that changes guanine for an adenine (G/A), was considered deleterious in 5 (83.33%) tools evaluated by PredictSNP2 (Table 1), but was not recognized by the GWAVA tool. This polymorphism causes amino acid substitution in the protein structure from serine (S) to phenylalanine (F) at position 512 (S512F), and was considered deleterious in all tools by PredictSNP1 (Table 2), indicating a possible alteration in protein structure and function. The MuPRO tool found a decrease in protein stability (ΔΔG = - 0.52385883). In addition, the Dynamut tool observed a decrease in the flexibility of the protein (ΔΔSVib ENCoM: -0.434 kcal.mol-1. K-1) and a change in the binding profile with adjacent amino acids due to the appearance of hydrogen bonds (mediated or not by water molecules), ionic bonds and hydrophobic contacts (Figure 1B).
In addition, the rs373441149 (C/T; G50S), rs145744844 (G/A; R51W) rs144452810 (C/T; D540N), rs372980300 (A/G; V632A) and rs1803711 (G/A; H754Y) polymorphisms are exonic and were considered deleterious in all the tools by PredictSNP2 (Table 1), except for the GWAVA tool, which did not identify these alterations. Concerning the analysis carried out by PredictSNP1 (Table 2), the polymorphisms mentioned above were considered deleterious in all the tools, except PANTHER. These modifications indicate important alterations in the DNA, as well as alterations in the structure and function of the protein. Finally, the rs201437141 (C/A; G39V), rs373145975 (C/T; V743M), and rs202065635 (A/G; I726T) polymorphisms are exonic. In the analysis carried out by PredictSNP2 (Table 1), they were considered deleterious in 5 (83.33%) tools, except for the GWAVA tool. rs377171913 (T/C; Y241C) was considered deleterious in 4 tools (66.67%), and neutral in the CADD tool (16.67%); the GWAVA tool did not recognize this alteration. rs201437141 (C/A; G39V) was considered neutral in PolyPhen-1 (12.50%), while rs373145975 (C/T; V743M) was considered neutral in PhD-SNP (12.50%) and both were considered deleterious in 6 (75.00%) tools by PredictSNP1 (Table 2); the PANTHER tool was unable to identify these alterations. However, rs377171913 (T/C; Y241C) was considered neutral in MAPP (12.50%) and rs202065635 (A/G; I726T) was considered neutral in PANTHER (12.50%), both being considered deleterious in 7 (87.50%) tools evaluated by PredictSNP1 (Table 2).
The NEDD9 protein is part of the CAS family of adhesion anchoring proteins [6] and is responsible for complexes of oncogenic kinases, focal adhesion kinases (FAK), regulating the duration of cell signaling cascades, controlling various processes of tumorigenesis and metastasis [10]. Its overexpression is related to a worse prognosis and it acts as an important physiological modifier in various types of cancer [10] such as breast cancer [11,12], lung cancer [13,14], prostate cancer [15], colorectal cancer [16,17], kidney cancer [18] and leukemia, especially AML [6]. Although NEDD9 is not considered an oncogene, research carried out on mice has confirmed its involvement in carcinogenic processes [21]. In addition, several studies indicate that this protein acts to promote metastasis in various types of cancer, especially triple-negative breast cancer, by stimulating the epithelial-mesenchymal transition [11-22]. Yuan Miao, et al [13], by analyzing lung tissues using immunohistochemistry and immunostaining, showed that overexpression of NEDD9 is related to lymph node metastasis and worse prognosis in non- small cell lung cancer [13], as it induces epithelial-mesenchymal transition via FAK activation, promoting metastasis [14].
Similar results have been reported in patients with bone metastasis from prostate cancer [15]. In addition, overexpression of the NEDD9 protein has also been linked to advanced stage and shorter survival in colorectal cancer [16] and renal cancer [22], and is associated with characteristics of tumor aggressiveness and progression [16-22]. The meta-analysis carried out by Yang Gu, et al [23], evaluated data relating to the overall survival of 2,393 patients with solid tumors, such as lung cancer, breast cancer, prostate cancer, colon cancer, and non- melanoma skin cancer, and showed that the elevation of NEDD9 mostly showed lower overall survival, which indicates a worse prognosis and could be used as a potential biomarker and a new therapeutic target [23]. Shenghao Hua, et al [6], through in silico analysis using the TCGA database and the GEPIA server, identified the overexpression of NEDD9 in patients with AML. BloodSopt showed its high expression, especially with the FAB-M4/M5 subtypes, since overexpression of the NEDD9 protein is related to monocytic lineages [6]. Interestingly, the NEDD9 protein has opposite effects on myeloid cells, since it blocks the migration of neoplastic myeloid cells, while in solid tumors and lymphoid leukemias it stimulates cell proliferation [6-24].
In a complementary way, it was identified that patients with high expression of NEDD9 had a lower overall survival rate, which is considered a worse prognosis and could be used as a potential biomarker in AML [6]. The literature also reports that overexpression of the NEDD9 gene was related to a good prognosis concerning intermediate-risk acute myeloid leukemia. The gene expression of NEDD9 was evaluated in two cohorts of patients aged over 50 years old, diagnosed with AML- IR, and the analysis identified, for the first time, the overexpression of NEDD9 as a new biomarker of improved overall survival [24]. However, AML is a neoplasm that occurs mostly in patients of advanced age [6]. There have been several studies mentioning the high expression of the NEDD9 protein in several types of cancer [6-18], but concerning AML and its polymorphisms, the literature is still scarce.
Single nucleotide polymorphisms are genetic variations in a single nucleotide in the DNA sequence and can be in regions that code for the protein, generating amino acid changes in the protein structure [25]. Polymorphisms can explain phenotypic variations between humans and are useful for explaining susceptibility to various diseases [25]. In recent years, data- theoretic and analytical methods, modeling and computer simulation techniques for biological studies have been widely used to simulate gene predictions, define structural and physicochemical properties of proteins, phylogenetic analyses, and simulate the interaction of biomolecules in a living cell, thus aiding in studies [26]. Bioinformatics uses computational methods and biological research tools and is advantageous for predicting the structure, function, and evolution of unknown genes and proteins. It also helps in the development of vaccines and medicines [27] and plays a fundamental role in the analysis of polymorphisms present in the gene of interest [26].
In silico analysis of proteins and nucleotides has been increasingly used in the field of bioinformatics, helping significantly in studies of SNPs, due to its flexibility, low cost, speed, and high precision, being considered genetic markers [26]. Using a vast repertoire of bioinformatics tools, we found 12 polymorphisms capable of generating amino acid changes and altering protein function, stability, flexibility, and structure. We demonstrated that rs185882325 (exonic; T/C; Y261C) can alter the structure and function of DNA, and promote protein alterations, concerning function, structure, decreased stability, and increased flexibility of the protein. Similarly, rs368480197 (exonic; G/A; R449C) and rs371007711 (exonic; G/A; S512F) can alter the structure and function of the gene (PredictSNP2), reduce stability (MuPRO) and promote changes in the protein, as both were considered deleterious in all the tools evaluated by PredictSNP1. Also, rs368480197 was found to increase the protein's flexibility and modify its binding profile with adjacent amino acids, due to the appearance of halogen bonds (Dynamut). And rs371007711 (exonic; G/A; S512F) showed a decrease in protein flexibility and changes in the binding profile with adjacent amino acids due to the appearance of hydrogen bonds (mediated or not by water molecules), ionic bonds and hydrophobic contacts (Dynamut). Although the literature is currently scarce concerning the relation between NEDD9 polymorphisms and AML, our analysis showed that rs185882325, rs368480197, and rs371007711 of the NEDD9 gene could be used as possible candidates for biomarkers in several types of cancer including AML, which could be used as promising genetic biomarkers and could contribute to diagnosis, prognosis prediction, risk and even help as a therapeutic target.
The NEDD9 protein has an important role in the regulation of cell signaling cascades, controlling tumorigenesis and metastasis processes. Our analysis showed 12 polymorphisms that promote amino acid changes in protein structure. The rs185882325, rs368480197, and rs371007711 were considered capable of modifying the structure, function, and stability of NEDD9 protein and could play an important role in several types of cancer including AML, therefore, they might become important biomarkers and deserve further investigation in AML patients.
The authors thank the Department of Biomedicine of Nossa Senhora do Patrocínio University Center (CEUNSP), Itu/SP, Brazil.
Do not apply.
Concept: LTR, RPR, KGRC and LSB; Design: LTR, KGRC and LSB; Data Collection or Processing: KGRC, LSB and DZV; Analysis or Interpretation: LTR, RPR, KGRC, LSB and DZV; Literature Search: KGRC and LSB; Writing: LTR, KGRC, LSB and RPR.
The authors declared that study did not receive financial support.
The authors declared no conflict of interest.


