Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Anna Julia Piva, Juliana Dario, Davi Zanoni Valente, Regiane Priscila Ratti Sartori and Larissa Teodoro Rabi*
Received: August 29, 2024; Published: October 02,2024
*Corresponding author: Larissa Teodoro, Rabi Department of Biomedicine, Nossa Senhora do Patrocínio University Center (CEUNSP), Itu/SP, Brazi
DOI: 10.26717/BJSTR.2024.58.009219
Acute myeloid leukemia (AML) is characterized by abnormal proliferation of immature myeloid cells. Although it has no specific factor for development, it may be linked to genetic inheritance, lifestyle or exposure to toxicity. Bioinformatics analyses enable the screening and identification of diagnostic or prognostic biomarkers for AML. The ASXL1 gene is associated with hematopoietic cell differentiation, mutations and polymorphisms in the sequence that are considered risk factors for the development of AML, often associated with poor prognosis and low survival rate. The aim of this study was to identify the main structural, functional and protein stability modifications resulting from the presence of polymorphisms in the ASXL1 gene. This is an in-silico analysis performed using 10 free and open access bioinformatics tools (PredictSNP1.0, SIFT, PolyPhen-1, PolyPhen-2, MAPP, PhD-SNP, SNAP, PANTHER, MuPRO and DynaMut). A total of 175 polymorphisms in the ASXL1 gene were evaluated, of these 13 (7,42%) polymorphisms were considered deleterious in 7 tools, suggesting important alterations in protein structure.
The polymorphisms selected were: rs139716375 (I268F); rs140896392 (A1512T); rs142450571 (G704R); rs143594461 (C55R); rs145061712 (S767Y); rs146743741 (D1525N); rs147905623 (D1230Y); rs148144203 (R1318W); rs199681643 (H1524Y); rs200053121 (S304C); rs3171367 (G1520R); rs370054224 (V179M) and rs373418380 (R265H). Thus, the bioinformatics screening allowed the selection of 13 target polymorphisms for the development of new studies to understand their applicability as possible biomarkers of diagnosis, prognosis, risk or choice of more effective treatment in neoplastic diseases, mainly associated with cases of AML.
Descriptors: ASXL1; Leukemia Myeloid Acute; Polymorphism Genetic; Protein Stability
Acute myeloid leukemia (AML) occurs due to a failure in the bone marrow that directly affects hematopoiesis [1], as it is a heterogeneous malignancy resulting in a neoplasm of myeloid precursors, causing the release of blasts into the bloodstream and a leukemic infiltration in the bone marrow [2]. AML does not have a specific factor for its onset; however, there are some factors that may be linked to its development, such as genetic inheritance, individual habits that are harmful to health and lifestyle (e.g. smoking and alcoholism) and exposure to toxic substances that are harmful to human health, such as electromagnetic radiation emitted by some devices like X-rays [3,4]. According to the National Cancer Institute (INCA), leukemias are the tenth most common type of cancer. Between 2023 and 2025, there are an estimated 11,540 new cases per year. Its incidence in men is approximately 6,250 cases, while in women it is 5,290 cases per year.
In addition, in 2020 there were around 6,738 deaths from AML [5]. The diagnosis of AML can be made using various laboratory tests, such as the complete blood count (CBC), which is the first test to be requested in cases of suspicion, since it is a low-cost, non-invasive test capable of analyzing the patient's general condition, as well as making it possible to differentiate morphologically between healthy cells and neoplastic cells, such as the presence of > 20% blasts, which is generally reported by the equipment [4-7]. In addition, the clinical symptoms observed in these cases are diverse, so assessing the patient's clinical condition is extremely important [4]. The main signs and symptoms are: the appearance of purple spots on the body, feeling unwell, febrile neutropenia, anemia and intense and persistent leukocytosis, most often with the presence of myeloblasts [4-7]. In addition, other laboratory tests are also requested, mainly including: myelogram, fluorescence in situ hybridization (FISH), cytochemistry (MPO and Sudan Black), flow cytometry (CT) and immunophenotyping. These tests are more specific and capable of tracking the leukemic lineage of the blasts, as well as the different stages of maturation of the cells present in the patient's sample [4-11].
In addition to the tests mentioned above, identifying chromosomal abnormalities (CA) is extremely important in the diagnosis. This assessment is carried out by the medical cytogenetics department and makes it possible to observe and evaluate the morphological, structural and functional alterations present in the patient's chromosomes [12]. CA can be associated with the presence of mutations and/or genetic polymorphisms [13]. These genetic alterations can generally act as biomarkers for diagnosis, prognosis and even risk for the development of future diseases, so the use of bioinformatics databases helps in the process of research, evaluation and understanding of the possible clinical usefulness of these mutations and/or polymorphisms [14]. The literature reports the involvement of several genes in the development of AML [5]. The ASXL1 gene (Additional Sex Combs-Like), located on chromosome 20, locus q11, is expressed in all types of hematopoietic cells and has the function of encoding the protein of the same name, responsible for modifying the histones of chromatin [15,16]. ASXL1 is an important epigenetic regulator in the process of hematopoiesis and clonal expansion, as well as being essential for the differentiation of hematopoietic cells into lymphoid progenitors and myeloid progenitors [17]. In a study carried out in 2019, which used human and animal hematopoietic cells, specifically mice, it was observed that the ASXL1 gene, by eliminating its wild protein, is the main cause of inefficient hematopoiesis, weakening the process and accelerating the development of myeloid malignancies, increasing the risk of mortality.
Therefore, mutated expression of the ASXL1 gene together with a negative Philadelphia chromosome (PHD) is associated with a poor patient prognosis [15-17]. In addition, ASXL1 is part of the Enhancers of Trithorax (TrxG) and Polycomb (PcG) groups, which encode essential regulators for cell differentiation and maturation [18]. Proteins from the TrxG and PcG groups modulate gene expression by controlling the chromatin state modification complex, keeping it repressive or active at the target locus, respectively, and also the transcription factor during hematopoiesis [18]. It is also responsible for preserving the epigenetics of ASXL1 and maintaining the standardization of gene expression during cell division and replication [18]. Nevertheless, ASXL1 is one of the genes with the highest mutation rate in all subtypes of malignant myeloid neoplasms, and is therefore the gene with the highest mutational recurrence in clonal blood diseases characterized by ineffective hematopoiesis, cytopenia and dysplasia [19].
The main alterations in ASXL1 are caused by the deregulation of proteins with suppressor functions and are mainly related to the presence of mutations or genetic polymorphisms [20]. The AML1-ETO mutation in ASXL1 is often linked to the RUNX1 mutation, both of which come from the translocation of chromosome 8 with chromosome 21 (t8:21), resulting in effects correlated with the uncontrolled acceleration of leukomagenesis [18,7] Genetic polymorphisms are variations that can occur in the nucleotide sequence of DNA, such as the substitution, deletion or insertion of nitrogenous bases, which directly influences the chain of amino acids responsible for making up the coding protein of DNA.
Genetic polymorphisms are usually a type of consequence of mutations, being a slightly more frequent event when compared to the mutation itself. Polymorphisms usually appear in more than 1% of the population, while genetic mutations appear less frequently, around less than 1% [21]. In particular, missense polymorphisms are capable of altering the amino acid (AA) composition of the protein, since they occur in coding regions of the gene. This change in AA can lead to structural, functional and/or protein stability changes [22-25]. The aim of this study was therefore to identify the main polymorphic alterations in the ASXL1 gene that can influence protein structure, function and stability.
This is an in-silico analysis in which the main polymorphisms recorded for the ASLX1 gene were screened. The polymorphism information was obtained from the Polymorphism Database (dbSNP) of the National Center for Biotechnology Information (NCBI) [26]. The protein structure information was obtained from The Universal Protein Resource (UniProt) [27] database under the code "Q8IXJ9" to obtain the FASTA sequence of the ASXL1 protein. PredictSNP1 [28] was used to identify potential morphofunctional modifications to the protein resulting from the change in amino acids due to the mutation in the ASXL1 gene and its new ionic interactions. These changes could have an impact on the stability, flexibility and functionality of the protein. The site has eight analysis tools, including MAPP [29], which performs the physicochemical analysis of protein variations; Polyphen-1 [30] and Polyphen-2 [31] are responsible for the practical analysis of the physical properties of molecules, as well as structural impacts and protein functions; SIFT [32] processes estimate of amino acid substitutions, as well as analyzing their protein functions and chemical characteristics;
PANTHER [33] performs protein analysis; nsSNAnalyzer [34] evaluates phenotypic impacts; PhD-SNP [35] uses the Support Vector Machine methodology to evaluate protein structures and sequences; SNAP [36] is responsible for analyzing the protein's secondary structural alterations.PredictSNP2 [37] was used to detect polymorphonuclear gene alterations in relation to the effects of nucleotide substitution in the genetic sequence, the site provides six tools for the analysis; the CADD tool [38] prioritizes functional, deleterious and pathogenic variants in various functional categories, effect sizes and genetic architectures; the DANN tool [39] classifies, identifies and understands the pathogenicity relationships between genetic resources; the FAT tool [40] evaluates the functional impact of pathogenic variants obtained from the literature, extracted from the Human Gene Mutation Database (HGMD).
The FUN tool [41] uses an empirical scoring system integrating evolutionary constraints together with epigenetic data to assess the impact of variants; and the GWAVA tool [42] uses the same method as the FAT tool, but GWAVA was produced to analyze regulatory variants [42]. In addition, the MUPro [43] tool was used to analyze protein stability in terms of Gibbs energy, corresponding to the highest energy that the molecule is capable of transforming into work, equivalent to the possibility of polymorphic alterations. The DynaMut [44] tool was also used to model possible protein alterations and assess their impact on morphological and functional characteristics, such as protein stability and flexibility. Thus, these 16 tools provided a robust analysis of the morpho functional changes in the protein resulting from the presence of polymorphisms in the ASXL1 gene.
The polymorphisms of the ASXL1 gene were investigated between March/2023 and June/2023. We found 34,257 registered polymorphisms for the ASXL1 gene, of which 2,465 are missense polymorphisms, 175 of which are capable of promoting AA alterations in protein structure and are therefore the target of our study. The 175 polymorphisms selected promote AA alterations in protein structure, which were evaluated using the PredictSNP1.0 tool (Appendix 1). Of these, 13 polymorphisms (Table 1) were considered deleterious in at least 80% of the tools evaluated and were therefore evaluated by PredictSNP2.0 (Table 2) in an attempt to understand how these alterations could affect the structure of cellular DNA. It is also important to note that none of the alterations evaluated were recognized by the nsSNPanalyzer tool.
Table 1: Results of the in-silico analysis of amino acid changes caused by polymorphisms in the ASXL1 gene obtained using the PredictSNP1 tool.
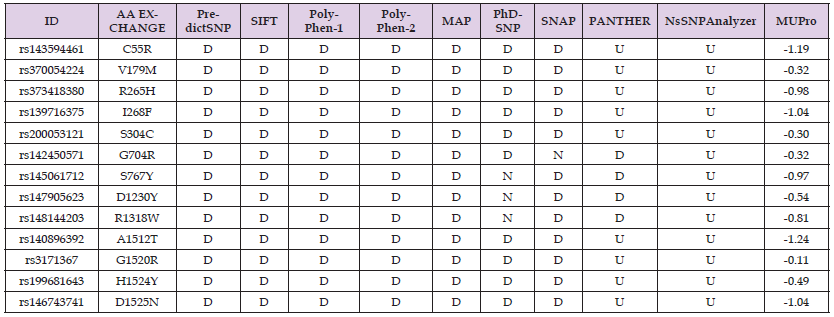
Note: Abbreviation: ID – SNP Identification; AA – Amino acid; D – Deleterious; N – Neutral e U – Unknown
Table 2: Result of the in-silico analysis of nucleotide changes resulting from the presence of polymorphisms in the ASXL1 gene obtained using the PredictSNP2 tool.

Note: Abbreviation: ID – SNP Identification; AA – Amino acid; D – Deleterious; N – Neutral e U – Unknown
The rs1435294461 (G/C; C55R), rs370054224 (C/T; V179M), rs373418380 (G/A; R265H), , rs139716375 (G/A; I268F), rs200053121 (A/T; S304C), rs140896392 (G/T; A1512T), rs3171367 (G/A; G1520R), rs199681643 (A/C; H1524Y), rs146743741 (G/C; D1525N) were evaluated by the PredictSNP1.0 tool (Table 1) and were considered deleterious in 7 of the 9 tools; however, the PANTHER and nsSNPanalyzer tools were unable to evaluate any of the polymorphisms mentioned. Thus, these 9 polymorphisms were considered deleterious in all the tools capable of evaluating them. They were also considered capable of reducing protein stability by the MuPRO tool. In contrast, the rs142450571 (G/A; G704R), rs145061712 (C/A; S767Y), rs147905623 (A/C; D1230Y) and rs148144203 (C/T; R1318W) polymorphisms, also assessed by the PredictSNP1. 0 (Table 1) were considered deleterious in 7 of the 9 tools used, with the rs142450571 (G/A; G704R) alteration being considered neutral only in the SNAP tool. The other changes resulting from the rs145061712 (C/A; S767Y), rs147905623 (A/C; D1230Y) and rs148144203 (C/T; R1318W) polymorphisms were considered neutral only in the PhD-SNP tool. Thus, these 4 polymorphisms were considered deleterious in 87.5% of all the tools capable of evaluating them. In addition, it was also observed that these alterations can lead to a decrease in protein stability.
In addition, rs370054224 (C/T; V179M), rs139716375 (G/A; I268F) and rs373418380 (G/A; R265H) were considered deleterious in 100% of PredictSNP2 tools. 0 (Table 2) and rs1435294461 (G/C; C55R), rs200053121 (A/T; S304C), rs142450571 (G/A; G704R), rs140896392 (G/T; A1512T), rs3171367 (G/A; G1520R), rs199681643 (A/C; H1524Y), rs146743741 (G/C; D1525N) were considered deleterious in 5 of the 6 tools evaluated, suggesting relevant impacts on DNA structure. The rs148144203 (C/T; R1318W) polymorphism was considered deleterious in only 1 of the 6 tools evaluated by PredictSNP2.0, while the rs145061712 (C/A; S767Y), rs147905623 (A/C; D1230Y) polymorphisms were not considered deleterious in any of the 6 tools used for evaluation in PredictSNP2.0. Therefore, they are considered polymorphisms that do not suggest relevant impacts on DNA structure and, consequently, on AA structure.It was observed that the C55R exchange (rs1435294461; G/C) promotes a change in the bonding pattern with adjacent AA, where in the wild-type structure of the protein there is the removal of a hydrogen bond in the lower portion, while in the altered structure, it was possible to observe the gain of a new ionic interaction in the central portion of the chain (Figure 1A).
Similarly, in the R265H exchange (rs373418380; G/A), in the wild-type structure it can be seen that hydrogen bonds have been lost in the lower central region and halogens in the upper portion of the right-hand end of the chain. In the altered structure, there was a gain of ydrogen bonds mediated by water in the upper and lower portions of the right lateral end, and a new carbonyl contact, also located in the upper region of the direct lateral end of the chain (Figure 1B). The rs200053121 (A/T; S304C) is also capable of altering the bonding pattern of the adjacent AA in the protein structure, where hydrogen bonds (mediated by water or not) are gained in the lower central portion of the chain (Figure 1C). In the D1230Y exchange (rs147905623; A/C) it is possible to observe the inclusion of new ionic interactions in the lower central portion of the chain, as well as the inclusion of a new halogen bond in the lower portion of the right side. Again, similarly, the G1520R exchange (rs3171367; G/A) removed ionic interactions and a hydrogen bond mediated by water. The rs199681643 (A/C; H1524Y) promotes the substitution of a histadine for a tyrosine at position 1524 of the protein chain.
This substitution is capable of modifying the interaction with adjacent amino acids, promoting the inclusion of new ionic interactions in the right lateral portion of the chain, as well as several new approximation structures, also located in the right lateral portion. As well as the exchange of AA resulting from the presence of rs146743741 (G/C; D1525N), which promotes the removal of hydrogen bonds mediated by water in the upper portion of the right side of the structure, and in the altered structure, it is possible to observe the inclusion of new ionic interactions in the central portion of the protein chain. In addition, the alteration resulting from the presence of rs139716375 (G/A; I268F), rs370054224 (C/T; V179M), rs142450571 (G/A; G704R), rs145061712 (C/A; S767Y), rs148144203 (C/T; R1318W) and rs140896392 (G/T; A1512T) did not show any alteration in the binding pattern with adjacent AA.
Genetic alterations that impact on hematopoietic function are becoming increasingly noticeable, mainly due to the growing importance of genetic tests in diagnostic processes and estimating prognosis and/or risk [45]. Alterations in the ASXL1 gene are frequently detected in various types of malignancies of the myeloid lineage, such as Aplastic Anemia [45], Myelodysplastic Syndromes (MDS) [45], Myeloproliferative Neoplasms [45], Chronic Myelomonocytic Leukemia and especially in cases of AML [45]. In addition, according to Fugino T et al. [46] the presence of ASXL1 alterations is related to poor prognosis in cases of hematopoietic diseases [46].The ASXL1 gene, which belongs to the ASXL [47] family, is located on chromosome 20, locus q11[15] and is capable of encoding the ASXL1 protein, belonging to the TrxG and PcG groups, which are responsible for modifying the histones of chromatin [16]. Mutations occurring in the Asx sex combs of the ASXL1 gene affect the homeotic phenotype characteristic of both groups belonging to it, such as PcG, the recessive complex, and TrxG, the activating complex, causing a phenotypic deficiency, according to Gao X et al [47].
In a complementary way, Fugino T et al. [46] point out that alterations in the TrxG and PgG groups, in fact, mainly affect the differentiation of cell lineage patterns [46]. Similarly, Mousa NO et al. [48] also found that the presence of mutations in ASXL1 related to the TrxG and PcG groups alters the ability to modify histones [48], and Medina EA et al. [18] point out that cell lineage patterns actively depend on the expressions made by the histones that have been altered along with their group [18]. Gao X et al. [47] and Yang FC et al. [49] report that the expression of the ASXL1 gene is constant throughout the embryogenesis process and subsequently in all adult tissues. Due to this constancy, it is part of the list of genes that are often most affected in clonal hematopoiesis of undetermined potential (CHIP), along with the RUNX1, NRASG12D, TET2 and DNMT3A genes. CHIP is strongly related to ageing and smoking, increasing the risk of developing malignant myeloid diseases by up to 10 times [47-49]. Furthermore, according to a gene expression study carried out by Mousa NO et al. [48], a 52% increase in ASXL1 expression levels can be observed in patients with AML, reiterating its correlation with hematopoietic processes and the worse prognosis of patients affected by the disease [48].
According to Asada A et al. [50] CHIP is characterized by a clonal expansion of pre-leukaemic somatic mutations in the absence of evidence of hematological malignancies, and is also associated with a high risk of developing neoplasms, as well as being related to poor prognosis in relation to haematopoietic diseases [50]. Asada A et al. [51] point out that ASXL1 alterations have a negative impact on AML cases, reiterating that ASXL1 is necessary for the silencing of phenotypically deficient and mutant hematopoietic genes [51]. Such mutations affect individuals with clonal hematopoiesis of undetermined potential (CHIP), Rahmani ZNE et al [52] state that ASXL1 mutations are more frequent in these patients, due to the direct alteration in PcG proteins. In addition to clonal hematopoiesis of undetermined potential, Montalban-Bravo LG et al [53] point out that ASXL1 is also linked to secondary AML.
Although the literature is extensive in relation to gene expression and the consequences of mutations in ASXL1 [52,53] there are few articles that report the presence of genetic polymorphisms, especially in missense regions and that promote alterations in the amino acids of the protein structure. Therefore, our study is a pioneer in the in-silico identification of nine polymorphisms with MAF greater than 0.1 that may be correlated with the development and even the pathophysiological processes associated with AML. The changes in proteins caused by the presence of these genetic polymorphisms may result in structural, functional or protein stability modifications. Recent biological analyses have also shown that mutant ASXL1 plays crucial roles in leukemogenesis and increases susceptibility to myeloid transformation by altering histone modifications. [46,47]
Fugino T et al. [46] reiterates that alterations in ASXL1 actively affect the differentiation of cells in accordance with normality, since these alterations can modify the protein function of ASXL1, causing loss of function and impairment of the metabolic processes in which it is involved [46]. This highlights the importance of evaluating structural, functional and protein stability aspects. Our analysis showed that the polymorphisms rs139716375 (I268F); rs140896392 (A1512T); rs142450571 (G704R); rs143594461 (C55R); rs145061712 (S767Y); rs146743741 (D1525N); rs147905623 (D1230Y); rs148144203 (R1318W); rs199681643 (H1524Y); rs200053121 (S304C); rs3171367 (G1520R); rs370054224 (V179M) and rs373418380 (R265H) are able to modify the structure, function and stability of ASXL1.
There are also increasingly clear indications that the natural presence of the mutant ASXL1 protein disrupts the process of blood cell formation and stimulates the change to a malignant form of myeloid cells, altering modifications in histones in a way that inhibits normal function and, at the same time, activates gain-of-function mechanisms [54,55]. In addition, new and promising therapeutic strategies are being explored to effectively treat hematopoietic malignancies that have alterations in the ASXL1 [56-58] gene. Bewersdorf JP et al. [59] reported that the presence of alterations in ASXL1 promotes an increase in the response rate to therapy using the Anti-PD1 antibody Nivolumab, but additional immunophenotypic studies are still needed to confirm these findings.
The ASXL1 gene is responsible for encoding the ASXL1 protein, which plays an epigenetic preservation role during the hematopoiesis process, in order to maintain the stability and standardization of histones during clonal expansion, with a view to cell suppression. The consequences of the presence of polymorphisms in the ASXL1 gene in missense positions with the capacity to alter amino acids in the protein structure have not yet been well elucidated in the literature. Our data show that after evaluating 175 missense polymorphisms, 13 [rs139716375 (I268F); rs140896392 (A1512T); rs142450571 (G704R); rs143594461 (C55R); rs145061712 (S767Y); rs146743741 (D1525N); rs147905623 (D1230Y); rs148144203 (R1318W); rs199681643 (H1524Y); rs200053121 (S304C); rs3171367 (G1520R); rs370054224 (V179M) and rs373418380 (R265H)] have MAF greater than 0. 1 and were considered deleterious in at least 80% of the tools used, so they may become important biomarkers in clinical practice and deserve further investigation in AML patients.
No conflict of interest was declared by the authors.
The authors declared that study received no financial support.


