Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Bright E Igere*
Received: August 23, 2024; Published: September 13, 2024
*Corresponding author: Bright E Igere, Microbiology Unit, Department of Biological Sciences, Dennis Osadebay University Asaba, Delta State, Nigeria and Biotechnology and Emerging Environmental Infections Pathogens Research Group (BEEIPREG), Microbiology Unit, Department of Biological Sciences, Dennis Osadebay University, Asaba, Delta State, Nigeria
DOI: 10.26717/BJSTR.2024.58.009176
Considering the rapid changing pattern, re-emergence and the global impact of the cholera pathogen in area of previously reported outbreaks, the problem of proper management and control/treatment remains a major concern. The profile of change at successive outbreak has become public health point of discuss in both developed and developing countries including Africa, Asia, Europe and Australia between 1992 till date. This review aimed to emphasize on the nature and relevance of such re-occurring variant in the management of impending outbreaks. Possible turning point to this discuss may be a redirected or a global re-evaluation of the identification/characterisation scheme, adequate preparedness for outbreak and global comparative genomic studies. An improved, developed and continuous epidemiological surveillance strategy that includes stringent application of molecular biology techniques on characterisation of transitional strains within countries and continents are suggested. These include bio-typing, sero-typing, sero-grouping, patho-typing, phage typing, plasmid profiling, mobilome typing, restriction fragment length polymophysm (Pulso-typing), typing of Cluster of Regularly Interspaced Short Palindromic Repeat (CRISPR) and the associated CRISPR (Cas) genes, typing of phage-inducible chromosomal island (PICI) or PICI-like element (PLE) typing, and antibacterial resistant pattern.
It is suggestive that future emphasis on improved comparative genomic studies, detection and identification/ characterisation scheme may influence control of outbreak and possible re-emergence problem. Application of the suggested scheme on early suspected cases both at the poor, rich, rural, urban, local and global sectors may affect the exact management and control of possible future outbreaks. With emphasis on global comparative genomics and perk-up characterisation scheme, the control/management of cholera or distribution dynamics as well as nature of re-occurring variant choleragenic Vibrio cholera, O1/O139 and Non-O1/O139 sero-groups there is high chance of effective control of strains.
Keywords: Vibrio Cholerae; Plasmids; Phage; PICI; PLEs; Choleragenic; Pathotyping; CRISPR; CAS and Pulsotyping.
Vibrio cholerae (O1/O139 and Non-O1/Non-O139) has continued to cause global food and water related diseases in Africa, Asia, Europe, America and Australia (WHO, [1]; Levade, et al.). In 2015, global reports shows that 42% occurred in Africa, 37% occurred in Asia while the remaining 22% occurred in Europe, America, Australia and other parts of the world. However WHO [1,2] in 2016, documented a total of 132,121 cases from 38 countries of the world, with 1.83% (2420) case fatality (death recorded). The reports of countries within the continent and percentage death shows Africa (17; 44.7%), Asian (12; 31.6%), European (4; 10.5%), America (4; 10.5%) and (1; 2.6%) in other countries. The later indicates that bulk of reported cases occurred in both Africa and Asia WHO [1,2]. Between 1994 and 2016, there had been an epidemic surge (eg the case of Somalia and Yemen) within the sub-Saharan Africa and Asia continent. The pathogen (7PET) was shown to be implicated in diseases and it possess/express varying pathogenic potentials, which necessitate a global surveillance intervention and comprehensive preventive/control programs (WHO [1]; Clemens, et al. 2017). Major virulence and resistance determining factors amongst Vibrio members tends to exhibit variability in expression at every successive outbreak. Earlier genomic sequencing studies of Colwell (2004) reported such variabilities as the pathogen transfers from one patient to another changing its virulent determining factors in successive outbreaks or infections. These variabilities and resistance determining factors are further confirmed in numerous phylogenetic studies (Colwell, 2004; Weill, et al. 2017; Igere, et al. [3,4]).
It was reported by various investigators that a wave of three sub-lineage which are genetically traced to a common origin were isolated from a collection between 1970 and 2014 (Didelot, et al. 2015; Weill, et al. 2017). The pathogenic deviance of seventh pandemic ET or strains (7PETs) and dynamism in cholerae studies produce overlapping gene sequences which are confirmed using other genetic or molecular typing approaches (Rebaudet, et al. 2013a, 2013b; Mutreja, et al. [5]). Predominant among the observed variabilities are the choleragenic toxin or cholera toxin (CT) (which mediates watery or rice stool resulting loss of fluid etc and today has become ctxA, ctxB and ctxAB) in an infection case and its co-regulator; the toxin co-regulated pilus (TCP) (Mutreja, et al. [5]; Didelot, et al. 2015). Beside pathogenic expression of the duo synergistically, other virulence factor (such as the hemolysin, RTX toxin, neuraminidase (nanH), mannose sensitive haemagglutinin (MSHA), cep gene for adhension, mannose-fructose resistance cell associated hemagglutinin (MFRHA) etc,) have also been implicated in Vibrio pathogenesis, re-emergence and environmental survival (Kaper, et al.[6]; Watnick et al. Chiavelli, et al. 2001; Faruque and Mekalanos, 2003a; Faruque, et al. [7,8]; Mandal, et al.[9]; Weill, et al. ; Igere, et al. [3,4,10]).
The survival mechanism of non-agglutinable Vibrio strains and horizontal gene transfer of virulent genes are tagged as contributing factors to their evolution, variability in pathogenesis in any of its successive re-emergence (Clemens, et al. 2017; Faruque and Mekalanos, 2003a; Faruque, et al. 1998b). Understanding and explaining the evolutionary event that portent the variable nature of V. cholerae at successive outbreak from choleragenic/non-choleragenic Vibrio is eminent. A specific recognition and study of transitional strains with incomplete related gene locus and function must not be avoided. It is presumed that such understanding would reveal the origin and reoccurrence of pathogenic strains especially the non-agglutinating serogroups. The study of Lu, et al. (2014), Faruque and his group (Faruque, et al. [7]) has revealed that non-O1/O139 environmental strain were carriers of few virulence associated genes. A similar previous study of Chakraborty, et al. (2000) on aquatic environment of cholera endemic region also shows the presence of diverse strains with various virulent gene profiles indicating that non- O1/O139 in aquatic regions are reservoirs of virulence genes. Such genes are presumed to have been transferred horizontally into O1/O139 making strains that were previously non-pathogenic to become pathogenic (Chakraborty, et al. (2000); Mukhopadhyay, et al. (2001); Lu, et al. (2014)).
Other similar studies also reported in addition that present in the genome of some Vibrio members are temperate or virulent phage which may be lysogenic. The lysogenic phage CTXΦ is a variant CT encoding gene dubbed ctxAB genes, whereas genes encoding toxin co-regulated pilus (TCP), which is responsible for major tissue colonization, are part of the TCP pathogenicity island (Kovach, et al. (1996); Waldor and Mekalanos (1996); Faruque and Mekalanos, 2003a; Clemens, et al., 2017). In addition, transmissible elements or self transposable elements (STE) (mobile element) were also found in association with some resistant gene markers amongst Vibrio members (Faruque, et al. [11,12]; Waldor, et al. 1996; Clemens, et al., 2017). Phage interaction with mobile transmissible elements, genes encoding the TCP pathogenicity island, the vibrio seventh pandemic island (VSP-1, VSP-2) and type III secretion system (TTSS) has resulted gene clustering with formation of new clone are not also left out in the discuss. They are shown to be possible major contributors to the emergence of new pathogenic strains (Mitra, et al. [13-15]; Faruque and Mekalanos, 2003a). From the forgoing, it is clear that the variability and resistant determinants observed amongst O1/O139 and non-O1/ non-O139 choleragenic V. cholerae in successive outbreaks has not been specifically traced to a single source factor. Hence the debate on the origin of variant strain remains contentious. This study is aimed at reviewing relevant reports on the variability’s in pathogenicity and antimicrobial determinants amongst O1/O139 and non-O1/ non-O139 choleragenic V. cholerae. It will also access observed/ documented reports in different endemic regions of the world over the last 20 years and the role of total/complete identificative characterisation scheme as a roadmap to the management and control of future impending outbreaks.
A systematically structured review of required literatures on spread and transmission of cholera cases in Southern African countries were collected from Web of Science, google scholar, Scopus, and Pubmed. Collected data includes environmental and clinical reports on variability of Vibrio strains, serotype and group.
The search engines were sourced between 14/06/2019; 12.06 GMT+2. The study adopted a title specific search following the with Boolean structure previously described by related investigators (Ekundayo, et al. [3,3,10,16-18]). The search also included molecular typing strategies, virulent genes, antibiotic resistant genes, etc).
All published documents were focus on cases of Cholera implicated by V. cholerae O1/O139 and Non-O1/Non-O139 in both clinical and environmental nexus while other non-conforming articles were excluded.
All collected articles were studied while relevant informations were collated and presented in figures and tables chronologically (Igere, et al. [18]).
More than 200 sero-groups of V cholerae have been identified with the somatic antigenic classification scheme to be implicated causal agent of most epidemic outbreaks of cholera (Igbinosa, et al. [18-22]; Clemens, et al. 2017). Those epidermic implicated somatic antigenic types are the O1 and O139, while others are refered to as the non-agglutinating (NAG) or somatic antigen absent types (SAAT). The basic classification scheme of choleragenic Vibrio member is divided in to the following: Serogroup, Biotype, Serotype, phage type, pathotype etc. Other classification schemes includes but not limited to bacterial growth/enumeration, biochemical identification, sugar fermentation, mobilome typing, antibiotic profiling, plasmid profile/type, antibiotic resistant marker and ribotypes (Igbinosa, et al. [19,23]). Other members of the somatic antigen determinants are O155, O129 etc. They are classified based on the presence of a bacterial outer membrane lipopolysaccharide (Shimada, et al. [24]).
The known biotypes of the choleragenic Vibrio members are the Classical and EI Tor, although some strains are referred to as Bengal strain and Calcutta strain because of their first place of detection and possession of additional characteristics (Mandal, et al. 2011). There are three main serotypes which have been associated with outbreaks; these are Inaba, Ogawa and Hikojima. The serotype classification is based on their possession of an antigenic component by the bacterium. These components are divided into A, B and C where B and C are yet to be characterised as at the time of collating data, A is a 3-deoxy- L-glycerotetronic acid. The Inaba possess A and C antigenic determinants, Ogawa possess A and B antigenic determinants while Hikojima possess all three (A, B, C) antigenic determinants. The absence of any of these antigens separates the organism as a non-serotype of the known serotypes (Shimada, et al. 1994; Uma, et al. 2003; Igbinosa, et al. [19]). Phage typing differentiates the Vibrio members into lysogenic phage type or non-lysogenic phage type (Summer, 2012).
It was observed by Faruque et al. [7] that various pathogenic characters in some strains of Vibrio are induced by lysogenic phage CTXΦ in an in-vitro induction process. There is high possibility that some non-identified environmental dynamics aids an induction process between the lysogenic phage and the V. cholerae pathogenicity. Hence phage particle CTXΦ is release into the environment as the phage utilizes a toxin co-regulated pilus (TCP) as its regulon region (Shlezinger et al., 2016). This is principal to the reason why lysogenic phage are able to infect Vibrio members that expresses TCP genes (Chakraborty, et al. 2000; Faruque, et al. [7]; Gorski, et al. 2015; Kutateladze, 2015). It is pertinent to note that the TCP genes are mostly harboured by O1 and O139 V. cholera which was also previously reported to have been associated with CTX genes (Gu, et al. [25]).
According to Igbinosa, et al. [19] and Kesik-Szeloch, et al. (2013), the assumption that phage’s are originators of variant formation is not farfetched as conversion by phage into novel strain ensures pathogenic character expression and variability among clones. The pathotyping scheme is employed to differentiate the various pathogenic strains and the pathogenic characters in an environment or a cholera endemic area, using the serenin (guinea pig eye test), rabbit ilea loop tests and Vero cells infection (Dziejman, et al. [15]; Begum, et al. 2006; Abia, et al. 2017). Currently, the understanding of gene ecology and application of molecular biology techniques has added the use of Polymerase Chain Reaction (PCR) in the detection of Cholera Toxin genes, TCP genes and other virulence or pathogenesis related genes (Mitra, et al. [26,27]; Mukhopadhay, et al. 2001; Abia, et al. 2017). Nonetheless, other recent advances in the metabolic activities of pathogen and interest in high throw-put detection and characterisation techniques has necessitated a stringent classification scheme for pathogens. Technological advancement and scientific mechanisation were not left out of the improvement recorded, as interest is now directed towards methods of high sensitivity and specificity employing the Matrix Assisted Laser Disorption Ionisation Time of Flight Mass Spectrometry (MALDI-TOF-MS). Application of this technology has made the characterisation of Vibrio members specific and timely. There is hope that in the near future, relevant high throw-put techniques would applied in patho-typing, phage-typing and other related typing schemes for the Vibrio pathogen.
Investigators in Vibrio studies have pointed out that haemolysins are dominant amongst the virulence factors of Vibrio parahaemolyticus and other members of the Vibrio family (Hao, et al. 2015; Fri, et al. [27]). These virulence factors are categorised into three major haemolysin gene types namely: thermo-labile haemolysin gene (tl), thermo-stable direct haemolysin gene (tdh) which is species-specific for V. parahaemolyticus, and thermo-stable-related haemolysin gene (trh) (Fri, et al. [27]). These haemolysin encoded genes (tl, tdh and trh) are currently applied in the pathotyping of both clinical, food related and environmental isolates of Vibrio members. (Bej, et al. 1999; Davis, et al. [27,28]; Nordstrom, et al. 2007). However, there is the inability to detect such toxin related genes using traditional microbiological techniques or the direct fluorescent antibody assay (DFA) in the field. In the early out break period of V. cholerae, typing systems were limited and not readily available (Faruque, et al. 1998a), making the detection and application of standard techniques labour intensive. Advances in detection techniques and research driven interest in molecular microbial genetics opened a new era for the detection of V. cholerae applying PCR techniques. This detection method was first applied by Bej, et al. (1999) using multiplex PCR technique which produced different band sizes (tdh: 269 bp and trh: 500 bp respectively). It was reported by Espineira, et al. [29] that some Vibrio members possess either tdh, trh or both genes at a time which probably may be responsible for the difficulty in treatment of the pathogen and possible variability in existence. A continuous study on virulence associated genes amongst the Vibrio members revealed other toxin genes.
Some of these toxin associated genes consists of ctx (cholera toxin), cep (core encoded pilin), zot (zonula occludens), ace (accessory cholera enterotoxin), pIIICTX (cholera associated), and psh or orfU (protease associated), tcp (toxin co-regulated pilus), rpo (DNA directed RNA polymerase) etc (Shah, et al. [30]; Jaiswal, et al. 2013; Yen, et al. 2017). These genes, all contribute to the morphogenesis and pathogenesis of the choleragenic Vibrio members. Vibrio strains that harbours virulent/pathogenic genes carry in addition, a prophage with cholera toxin known as CTXΦ (Kaper, et al. [6]; Plaza, et al., 2018). When the cells of pathogen finds itself in the host intestinal mucosa, it source for the enterocytes to anchor, while its released cholera toxin (CT) recognises and specifically binds to a GM1 receptor, resulting fluid accumulation, imbalance electrolyte transport system and increase/high influx of cyclic adenosine monophosphate (cAMP). The accumulated fluids are finally lost in the form of watery stool or profused diarrhoea (Broeck, et al. 2007). Hence, the dominant factor to any outbreak case in a pathogen is disease production by a virulent Vibrio member as indicated by the genomic sequence study on clinical V. cholerae strain conducted by Osama and colleagues (Osama, et al. 2012).
This is reported after an outbreak episode in Malaysia which shows the presence of multiple toxigenic genes (Osama, et al. 2012). The prophage (CTXΦ), is made up of two functional distinct clusters namely the RS2 and the core gene regions. The RS2 region encodes rstA, rstB and rstR which are responsible for recombination of the lysogenic phage to integrate onto specific regions of the bacterium. Phage replication in a host cell is achieved by rstA gene, integration by the rstB gene, and rstR for regulation of the prophage (CTXΦ) expression and/or repressor of rstA in any host or environment (Waldor and Mekalanos, 1996; Waldor, et al. 1997; Shah, et al. 2012). It is also reported that some member RS2 contribute to bacterial colonisation, phage replication and phage regulation. During this period, the rstR intergenic regions (ig-1 and ig-2) interact with the rstA to repress replication activity when the need arises (Shah, et al. 2012; Kimsey and Waldor, 1997). Result of previous studies has also shown that the rstR genes are biotype specific. It exhibits a heteroimmunity phenomenon that makes the EI Tor members to resist super-infectivity with similar biotype derived phage (EI Tor prophage) but accept the classical prophage rstR and integration (Kimsey and Waldor, 1997; Shah, et al. 2012; Sarkar, et al. [31]). The core consists of prophage morphological dynamics. A study on genomic characterisation of prophage shows that the core consists of ctxAB, cep, zot, ace, pIIICTX, and psh or orfU (Shah, et al. 2012; Jaiswal, et al. 2013; Yen, et al. 2017). These genes (zonula occludens (zot), core encoded pilin (cep), accessory cholera enterotoxin (ace), psh or orfU, pIIICTX and ctxAB all contribute to the morphogenesis and pathogenesis of phages and Vibrio members (Waldor, et al.1997; Shah, et al. 2012; Rajpara, et al. [32]).
Vibrio cholerae members have been found as resident flora of the environment before becoming pathogenic. The earlier studies John Snow 1853 and the Colwell (2004) substantiate that V. cholerae are found in the environment about fifty toxigenic genes and two chromosomes (the large harbours all the toxin genes). It is speculated that harsh environmental factors and phage conversion are contributing factors to the development of pathogenic strains. As it continue in being expose to tough environment, it acquire or take up new genetic material by horizontal gene transfer into their chromosome making a previous non-pathogenic organism to exhibit vital pathogenic characteristics (Faruque and Mekalanos, [33,34]). Such acquired character merits their extraordinary survival mechanism in harsh carbon sources (eg; chitin) (Meibom, et al. 2005). Another probable origin to emergence of new pathogenic strains is the addition and/or deletion of genes to existing chromosomal regions.
It was observed and reported in earlier studies of O139 sero- group that pathogenic clones transfer genetic element around the O-antigenic region which replaces lipopolysaccharide (LPS) O-antigen synthesizing enzyme (Bik, et al. [35]; Waldor, et al. 1994; Clemens, et al. 2017). In another study, it was observed that the presence of an integron element and a capsule polysaccharide in their outer membrane contributes to their pathogenicity (Faruque et al., 2003; Mandal, et al., 2011). However the evolution of pathogenic strain remains unclear amongst O1/O139 and non-O1/non-O139 strains (Faruque, et al. 2003b), yet the pathogenicity and epidemic cases continues to increase (Andrews and Basu, 2011; Bik, et al. [35]). The strains of USA Gulf Coast is not left in toxigenesis. Toxigenic gene found in the USA Gulf Coast (CTXUS Gulf) strains was unique to that region but similar to the classical strains as both possess rstRCla and ctxB1. (Kim, et al. [36]). Although few information’s are available on strains from Australia and other part of the world, there has been reports of numerous variants of CTX (CTX-1, CTX-3, -4, -5, -6, -7), and RS1 (rstREI Tor, rstRCla ), among strains from the seven pandemics. It is anticipated that newer pathogenic variant may be found in the eighth pandemic as well as newer CTX phages (Faruque, et al. 2007; Mutreja, et al. [5]). The surge is becoming unpredictable as it’s estimated globally that 1.3 – 4.0 million cases were reported of which over 21000-143000 deaths are being recorded per year (WHO, [2]).
The emergence of O139 choleragenic V. cholerae was trace to a previous occurrence of O1 biotype which occurred in the Asian continent. It was said to have co-existed with the EI Tor members found in the EI Tor village of Egypt (Harris, et al. 2011; Grim, et al. 2010). Being influenced by seasonal variation, a dual outbreak resulted with epidemic caused by two serogroup members dubbed O1 and O139 (Basu, et al. 2000; Faruque, et al. 1995, 1997b; Jutla, et al. 2011). Initially, V. cholerae O139 strains displaced the El Tor strains in 1992 but it was later displaced by a variant strain of O1 EI Tor in 1994, which dominated until 1996 in the Asian continent (Faruque, et al. 1997a; Basu, et al. 2000; Jutla, et al. 2011). By the seventh month of that same year, a new variant of O139 sero-group has emerged with new genetic character. It thrived in Calcutta with dominant expression of pathogenic dynamic until September 1997. While this was happening, between 1994 - 1995 reports from Central and Northern Bangladesh also shows a similar situation were there is displacement of O139 by O1 EI Tor strain.
The O139 strains continue to thrive in South Coastal region expressing their pathogenicity amongst man and the environment (Faruque, et al. 1997b, 1999; Basu, et al. 2000; Abd El Ghany, et al. 2014; Garrine, et al. 2017). Bangladesh also reported the occurrence of the O1 EI Tor member in 1996 with few O139 serogroup. Advances in technology and application of molecular biology techniques in detection of various Vibrio clones further aided the characterisation of variant strains, their toxin genes, related pathogenic genes and new genes of pathogenic relevance in the world. These advances also aided the differentiation of the US Gulf Coast strain from those of the seventh pandemic outbreak (Davies, et al.2017; Domman, et al. 2017; Clemens, et al. 2017; Basu, et al. 2000; Faruque, et al. [37] 1992, 1993, 1995; Wachsmuth, et al. [38], 1994; Waldor and Mekalanos, [39]). Further genetic analysis of strains from other parts of the world and those of the 1991 Latin American epidemic revealed a relationship with those of the 7PET outbreak; hence the strains were seen as the seventh pandemic extension (Weill, et al. 2017; Wachsmuth, et al. [38]; 1994; Domman, et al. 2017).
In a comparative study of the O1 EI Tor biotype and O139 strain from previous outbreaks, a striking relationship resulting from gene transfer was found indicating that the O139 clones originated from the O1 EI Tor members (Basu, et al., 2000; Faruque, et al. 1994; Wachsmuth, et al. 1994; Didelot, et al. 2015 Garrine et al. 2017). Similar reports were documented in a ribotyping study conducted by Faruque et al. (1995), with the non-O1/non-O139 members diverging wide from O1 and O139 strains on phylogenetic dendrogram. Other advanced studies on strains from previous outbreak (between 1961 - 1996) showed clonal differences, momentary gain and loss of six ribotypes among the classical biotype, presence of five ribotype among the EI Tor members and three ribotypes of O139 sero-group, presence of lysogenic phage and diverse CTX genotypes (Ali, et al. 2015; Basu, et al. 2000; Faruque, et al. 1993, 1994, 1995, 1996, 1997a, 2003b). The result of these studies also indicate a continuous re-emergence of new choleragenic clones through genetic succession by natural selection, environmental factors and host immunity (Basu, et al. 2000; Faruque, et al., 1995, 1996, 1997, 2003b; Abd El Ghany, et al. 2014; Garrine, et al. 2017).
Additional Reports on Choleragenic Vibrio Varabilities: The study of Faruque, et al. (2000) and his group reported that other toxin (choleragen) encoding gene acquisition occurs in some members of non-O1/non-O139 V. cholerae progeny which may be a new source of epidemic and pandemic clones (Faruque, et al. 1997, 2000). This has been confirmed by the study of Grim and his collegues (Grim, et al. 2010) who reported the occurrence of the seventh pandemic vibrio and detection of variant V. cholerae with variant gene acquired. Other recent studies on the non-agglutinating Vibrio cholerae member shows their implication in human systemic infection cases such as neonatal meningitis and septicemia (Lu, et al. 2014; Hao, et al. 2015). The choleragenic Vibrio cholerae of O139 serogroup is also shown to have originated from its recombination or sharing of genes with a choleragenic O1 strains (Faruque, et al. 2000). Most environmental studies on V. cholera strains had reported absence of the cholera toxin genes (Denner, et al. [40]), as well as the emergence of new choleragenic genes through possible acquisition of genetic material from the environment (Denner, et al. [40]). This describes the relevance of clinical management and control for the cholera toxin gene (ctxA) (Denner, et al. [40]). In an emergence and re-emergence study conducted by Faruque, et al. (1997) on V. cholera O1/O139 and non-O1/ non-O139 serogroup from four countries within Asia and Africa continents, A significant difference in the gene sequences flanking the pathogenic region was observed.
Their result also showed four different choleragenic (ctx) genotype and four dissimilar ribotypes. The result indicates an emergence of new clones of the El Tor vibrio strains which is different in pathogenicity from the earlier reported El Tor clones with similar structural, regulatory and co-regulatory pilus genes (tcpA, tcpI and toxR). In addition, these organisms are termed re-emerging since in any recent occurrence, a new pathological dynamics that threatens future disease situation is observed when compared to their previous occurrence. Some of such variations are possession of antibiotic resistant genes and/or choleragenic genes (Igbinosa, et al. [19,9]). Reports from several further studies demonstrate an appearance and disappearance of these toxigenic/choleragenic clones in India, Africa and European countries (Faruque, et al. 1995; 1993; Siddique, et al. [41,42]). Such reports show emergence and re-emergence of the epidemic Classical biotype strain and other non-O1 or O139 serogroups. It is pertinent to note that the V. cholera O1 of the El Tor biotypes was displaced by the O139 Bengal strains due to some possible undetermined environmental factors and host pre-existing immunity.
Suffice to say that the infection of the O139 Bengal Vibrio elicited an immune responds on the infected host as reported in recent study (Mandal, et al.[9]; Raheed, et al. 2013; Clemens, et al. 2017). The patient responds would include increased production of antibody secreting cells (ASC), vibriocidal agents and other antitoxic antibodies into their circulatory system. The immunoglobulin A (IgA), immunoglobulin M (IgM) and ASC response were stronger in O139 biotype compared to those in O1 biotype V. cholerae cases (Faruque, et al. [8]; Raheed, et al. 2013). Although their isotypes or magnitude of response to infection was similar, some striking differences were observed (Albert, et al. [43,44]; Raheed, et al. 2013). Other factors that ensure their infectivity are individual gastric acid barrier, blood group (high susceptibility amongst blood group O patients) and sanitary conditions (Clemens, et al. 1989; Barua and Paguio, 1977; Glass, et al. [45,46]). Suffice also to say that the Classical biotype which was the known causal agent of pandemic cholera (first to sixth pandemic) was later replaced by the El Tor biotype in 1961 with origin traceable to the Celebes Islands of Indonesia (Kaper, et al. [6]). After the displacement, between 1994 and 2005, the Vibrio O1 was shown to be less implicated in cholera cases (Alam, et al. [47]).
In 2001, it was documented that the detected Vibrio strain (7PET) showed a genetic change by harbouring a variant CT gene known as ctxB gene of the Classical biotype (ctxBCL) as the El Tor strain of similar gene (ctxBET) was said to be extinct (Alam, et al. [47]). This genetic change was first observed between 1998 and 1999 amongst O139 choleragenic V. cholerae (Bhuiyan, et al. [48]). Between 2010 and 2012, a surveillance study using both environmental and clinical isolates confirmed the presence of O1 (99.2%) and O139 Bengal strain (0.8%) with all isolates showing positive for ctxA, tlc, ace, zot and the El Tor biotype specific markers (hlyAET, tcpAET, and rtxC) (Rashed, et al. 2013). A ctxBCL allele (which is a variant of ctxBET) was also found in 98.4% of the total isolates of the V. cholerae O1 biotype El Tor strains. While 1.6% retained the parent gene ctxBET amongst the strains surveyed, with the variant strain having a nucleotide sequence different from the original allele. It was shown to carry a translated amino acid sequence with histidine replaced by threonine and tyrosine replaced by isoleucine at position 39 and 68 (Nair, et al. [49]; Rashed, et al.).
These observed variant strains were of high clinical relevance as they have been reported to be globally implicated in endemic cholera outbreak cases (Mutreja, et al., 2011). Other variability that occurred is the transient emergence of multiple-drug-resistant (MDR) strains (Glass, et al. [45]). It was observed in the study of Rashed, et al. (2013) that the ctxBET strain of O1 V. cholerae are closely related to previously reported 2001 strains. In the same manner, the ctxBET of O139 V. cholerae strains in previous studies of 1993 shows report of similar antibiotic resistant patterns as well as restriction enzyme digest band pattern using NorI. Other non-choleragenic or epidemic associated Vibrio species in halophilic environments include V. hollisae, V. alginolyticus, V. furnissii, V. metschnikovii and V. fluvialis. Although reports have shown that most Vibrio specie from marine environment including V. parahaemolyticus and V. vulnificus, are associated with most recorded outbreak cases (Harris, et al. 2012; Igbinosa, et al. [19,50- 54]]). Agreeably, different ecotypes have been reported in most epidemic outbreaks amongst the known serogroups (O1/O139) which either encode or does not encode any of CTX phage and TCP pathogenic dynamics. Although an extensive study revealed other pathogenic genes such as tcpA, tcpI, tcpH, acfB and CTX prophage (Hang, et al. 2003; Faruque, et al. 2004; Reen, et al. 2006; Schirmeister, et al. 2014), other genetic variant continue to resurface in any outbreak due to gene transfer mechanism. The strain of O139 which harbours a positive toxin co-regulatory protein gene undergoes a possible choleragenic conversion in the presence of a CTX phage during horizontal gene transfer (Faruque, et al. [33,55]).
V. Cholerae Phages, Virulent Genes and Pathogenesis Variabilities: After the emergence of the EI Tor Vibrio pathogenic strains, a further genomic characterisation study reveals the presence of an adjacent satellite phage called RS1, with similar genes (rstA1, rstB1 & rstR1) and character as the RS2 but differ by possessing an rstC gene which is an additional gene (Rajpara, et al. [32]; Shah, et al. 2012). The rstC encodes anti-repressor protein, which promotes transcription of phage (CTXΦ) genes and induction of ctxAB expression which is required for production of infectious particles (Rajpara, et al. [32]; Sarkar, et al. 2011). Suffix to say that this gives the higher virulence and choleragen produced by the EI Tor strains during the outbreak and the current nature of the cholera Vibrio (Rajpara, et al. [32]; Davis, et al. 2002; Waldor, et al. 1997). The adjacent satellite phage (CTXΦ) is a free replicative prophage which has demonstrated high genetic instability with increased pathogenic effect on V. cholerae (Jaiswal, et al. 2013). It possess two tyrosine recombinase genes (XerC and XerD) that mediate an irreversible CTX integration to ‘dif region’ (a specific attachment site in Vibrio chromosome) to produce increase virion copies using replicatory mechanism (Shah, et al. 2012; Davis, et al. 2002).
It was also demonstrated that during favourable conditions, stable lysogenic phages can be produced or propagated in toxigenic Vibrio strains that previously do not harbour phages (James, 2017; Jaiswal, et al. 2013). Such propagated phage’s may be integrated into the genome of the bacterial or remain as extra-chromosomal replicative phages. These are some of the activities of Vibrio phage’s that preclude the observed continous evolution of V. cholerae (Okada, et al. 2017; Sarkar, et al. 2011; Waldor and Makalanos, [39]; Faruque, et al. 1998a; Faruque et al., 1998b; Huber and Waldor, 2002; Val, et al. 2005; Das, et al. [56] 2010, 2011a, 2011b). Worthy of note is the establishment of the fact that previously non-toxigenic strains reemerge as toxigenic strains, as a result of phage infection and survival mechanism. Added to this is the co-evolution of genes arising from the transfer of genetic elements, and the affinitive recognition of TCP as potent receptor for propagation and/or infection by phages (Waldor and Makalanos, [39,57]; Faruque, et al. 1998a, 1998b,2002).
Diversity of El Tor Clones, tcp, toxR and choleragenic toxin gene ctx: The Vibrio biotype “El Tor” strains has been shown to exhibit some sought of clonal diversity amongst the population (Faruque, et al. [14,57,58]; Popovic, et al. 1993), suggesting the emergence of new clones from pre-existing parent. Colwell (2004), reported that V. cholerae diversity is confirmed fron the sequence data shown that the pathogen can inhabit various habitat. Faruque and Nair [37] and Merrell, et al. (2002) also observed a trend in the passage of V. cholerae amongst patient and reported that certain genes are lost due to transmission of cholera from patient to patient making the pathogen 700 times more infectious. There also reported a lateral transfer of genes and distribution of virulent genes amongst environmental strains of which provide an environmental reservoir for such genes giving credence to the diversity of thepathogen. The result of restriction digest study by Faruque, et al. (1997) on the O1 genomic 16S rRNA gene using the BglI endonuclease cleavage shows five different ribotypes.
The El Tor members of O139 Vibrios produces four ribotypes belonging to ribotypes I through IV, and 93.75% (30 of 32) belonging to ribotype V. This suggests a probable emergence of a new strain amongst the population of V. cholerae with the O139 members, having originated from the O1 strains (Kim, et al. 2015). A similar report has also been documented in other developed and developing countries from both environmental and clinical specimens. Their study also emphasized on the bacterial colonization of ilea region of the small intestine, as a crucial mechanism in V. cholera infections which is mediated by toxin-co-regulated pilus (TCP; subunits are tcpA, tcpH, tcpI) within the choleragenic toxin (CT) island and its regulatory milieus (ToxRToxT) (Ogierman, et al. [59]; Harkey, et al. 1994; Sarkat, et al. 1996; Taylor et al., 1987). Its restriction pattern in a restriction fragment length polymorphism experiment showed that these genes were different from the previously reported similar El Tor strains genes (Mutreja, et al. [5,56]). The difference is attributed to a phage expression of TCP using the CT as a receptor which suggests a possible bacteriophage lysogenic conversion using a gene called ctxF within the CT domain (Waldor and Mekalanos [39]; Kim, et al. 2015).
Hence some non-choleragenic strains are said to have arise from choleragenic strains and vice-versa. It also depicts a possibility of the emerging El Tor strain of O139 arising as a consequence of the bacteriophage lysogenic (or gene transfer and unfavourable environmental condition) activity on the non-choleragenic strain of pre-existing members or V. cholerae O1. In another restriction study of the CT domain, a different pattern of restriction digest was observed, suggestive of a possible integration of the phage at a particular site into the bacterial genome (Kendall, et al. 2010; Kim, et al. [36]). Such digest profile produced a 17-bp sequence called attRS1 in the non-choleragenic parent strain (Choi, et al. [60,61]). There is possibility that the new clone detected from environmental and clinical strains between 1994 to 1996 belong to the new ribotype I, while two other isolated strains from surface water environment between 1993 and 1994 belongs to ribotype II (Boyd, et al. [62]; Fazil, et al. 2011).
A similar result was observed in ctxA and zot genes encoding choleragenic toxin (ctxAB) and zonula occludens toxin (zot) (Naha, et al. [63]). In another perspective, there is a likelihood that present in the environment are low numbers of the new clone hence their detection was not positive. The report that at favourable conditions within the environment, strains begin to multiply making El Tor strains to thrive successively giving rise to a positive new strain (Albert, [64]; Kim, et al. 2015).
Biotype Variability
Unusual, Atypical and Dual Biotype Nature of Some Ei Tor Strain: The emergence and re-emergence of variant strains observed in the studies of various investigators have necessitated a debate on the biotype classification of the Vibrio cholerae strains. Recently, studies now begin to report unusual expressions of both classical and EI Tor biotype traits (dual biotype nature) by some V. cholerae strains (Zaw, et al. 2016; Plaza, et al. 2018). Some of them were also shown to harbour multiple pathogenic genes, variable antibacterial resistance, and presence of phage with diverse gene markers and plasmids. This observation was first reported by Nair and his group while working on O1 clinical isolates of Vibrio cholerae collected between 1991 and 1994 in Bangladesh (Nair, et al. 2002; Zaw, et al. 2016). Because these strains possess diverse dissimilar traits compared to the previously reported strains, they were called Bangladesh-Matlab variants. Safa and colleagues took further the study employing PCR based technique, pulse field gel electrophoresis and ribo-typing identification scheme on the RTX, VSP-I and VSP-II gene cluster of the EI Tor strains. Their result shows that the Bangladesh-Matlab variants originated from the EI Tor wild strain. Although there were diverse electrophoresis pattern similar to those of the classical biotype and EI Tor biotype traits, the variant was shown to harbour a ctxB gene similar to the classical phage cholera toxin (CTXΦCla) (Nair, et al. 2002; Safa, et al. [65], 2005; Zaw, et al. 2016).
After the Mozambique outbreak episode of cholera in 2004, studies on the causal agents (EI Tor strains) revealed that it possesses two additional traits (a small chromosome and two tandem copies of prophage). These traits were sequenced and identified as possessing similarity to the Classical phage (CTXΦCla) earlier detected in the Bangladesh-Matlab strains from the study of Safa, et al. [65] (2006) in both 2005 and 2006 (Das, et al. 2007; Faruque, et al. 2007). This was the first African based confirmation of dual biotype nature of these atypical variants also called ‘altered EI Tor strain’ with classical born phage (CTXΦCla), ctxBCla, ctxBEI Tor, and rstREI Tor. (Nair, et al., 2006; Nguyen, et al., 2009; Zaw, et al., 2016). Further studies of Nguyen and group revealed the presence of rstRCla and rstREI Tor genes in several strains indicating that these strains consist of two phage cholera toxin which occur as tandem or located in chromosome-1 and chromosome-2. Other studies in India on the O139 strains also showed similarity with the African atypical EI Tor strains (Das, et al. 2007). The group also reported the carriage of ctx-prophage in the small chromosome which encodes classical type ctxB (ctxBCla). A continuos study of such reported strains collected in Africa and Asian continent (some endemic countries were excepted) showed variant occurrence. Other similar characteristics were observed amongst previously reported strains in Bangladesh, India, Mozambique, between 1991-2004, as it was shown to harbor the ctxBCla, hence they were designated as ‘Hybrid EI Tor’(Safa, et al. 2008; Zaw, et al. 2016). The trend continues, the need for study is enforced, driving investigators to a suggestive positive eradication or control scheme.
Serotype Variability
Serogroup-Serotype and Variability: Following the serological identification scheme of V. cholerae, the sero-group is based on the possession of O-antigen (somatic antigen), dividing the members to O1/O139 and other non-O1/O139 (others are O129, O155 etc). However serotyping of Vibrio strains is based on possession of ABC antigenic character, which differentiate Vibrio members into Inaba, Ogawa and Hikojima (Igbinosa, et al. [19]). Studies on the O139 strain collected from the 1992 outbreak episode showed that these members (O139) were derived from an EI Tor descendant with a somatic antigenic (O-antigen) switch, which is probably facilitated by two genetic transfer activities - acquisition of phage by the bacterium and a transposable element associated with SXT (region of drug resistant to Sulfamethoxazole and Trimethoprim) (Bik, et al. [35,62]; Waldor, et al. 1997). This is particular to the characteristics observed amongst the Bengal and Calcutta O139 strains. Although the Bengal strain of O139 serogroup possess similar virulence factors as the O1 serogroup strains, there are differences between the two sero-members. These are possession/production of distinct O-antigenic trait and polysaccharide capsule by the Bengal O139 members which seem to be completely absent in the O1 serogroup members (Comstock, et al. [66]; Mooi and Bik, 1997; Stroeher, 1995).
These observations necessitated the suggestive statement of Bik, et al. [35] that the O139 members arose from a closely related strain of O1 EI Tor strain by acquisition of DNA. It was also traceable to insertion mutation at the somatic antigenic (O-antigen) region of the new strain (Mandal, et al. [9,67]). Further polymorphism and electrophoresis study on serotype by Bik and colleagues reveal that the O139 Bengal members show the presence of an otnAB (otnA and otnB) DNA which ensure their expression of a distinct cell surface antigenic property. The otnAB DNA was absent from the O1 strains but present in some non-O1 members (O69 and O141) necessitating their extra character (Bik, et al. [35]).
Case Fatality in Africa
Numerous reports from global investigators shows the upsurge of cholera cases in Africa with about 67% (417) of total global outbreak which is about 632 reported cases. Between 1995 and 2005, about 87.6% (423,904) cases were recorded amongst total global cases (484, 246) (Griffith, et al. 2006). In 2016 about 132, 121 cases were reported amongst 38 countries in the world, with 17 countries and 54% cases within the African continent involved. A total of two thousand and twenty deaths were recorded during this period with one thousand seven hundred and sixty-two deaths in Africa (WHO,[1]). Between 1991 and 1996, V. cholerae O1 EI Tor biotype has been implicated in a number of cases ranging from 70,000 to 160,000 which is the largest cholera case reported with 42% of death globally (WHO, [68]). Another cholera and shigellosis epidemic outbreak which occurred as a result of the Rwandan, Hutu and Tutsi tribal war, causing 12,000 deaths due to migration and living in makeshift camp in Goma and DR Congo was also implicated by the pathogen (Siddique, et al. [14,69]).
Although there were reported decline in the incidence and number of cases after the intervention of government, improved surveillance strategies serves as a major contributor to the decline (WHO, [1]). In December 1996 other reports from twenty-six countries within the African continent showed high case fatality with over 1000 cases in Democratic republic of Congo, Somalia, Nigeria, Senegal, etc (WHO, [70], [1]). A similar upsurge report was also reported between 2006 and 2010 (WHO [2,71,72]). De Magny, et al. (2012) and Emch, et al. (2008) pointed natural disaster and global weather change as the possible influencing factors of the upsurge of cholera, with Africa case fatality higher than 1.0%. The V. cholerae O1 serogroup is known for its increased virulence due to its high choleragin production (Ghosh-Banerjee, et al. 2010). Although there has been an upsurge of cholera reports in Africa, there are few reports on the occurrence of variant V. cholerae O1 El Tor. The included countries in Africa are Nigeria, Cameroon (Quilici, et al. 2010), Mozambique (Ansaruzzaman, et al. [73]), Angola (Ceccarelli, et al. [74]), South Africa (Ismail, et al.2012), Zimbabwe (Islam, et al. 2011), Zanzibar (Naha, et al. 2013), Kenya (Saidi, et al. [75]) and DR Congo (WHO, [1]).
Although the study of Kiiru, et al. (2009) failed to detect any variant strain, a similar study by Mohamed, et al. (2012) reported a classical bio-type with ctxB gene which is a variant of the CTX gene. The study of Shikanga, et al. (2009) and Saidi, et al. [75] on V. cholerae O1 El Tor variant showed a severe form of the strain as its disease manifestation was rapid. This manifestation necessitate these investigators to carry out an association or relationship study on strains collected from 2007-2008 and 2013 cholera outbreak with the variant form of V. cholerae O1 El Tor. They observed from their report that there was a high and most severe infectivity in all forms of the V. cholerae O1 El Tor strains from the cholera epidemic (Shikanga et al. 2009; Saidi, et al. [75]). A similar outbreak in Zimbabwe between August 2008 and February 2009 killed over 3800 persons (Kapp [76,77]). From the forgoing, it is evident that the variant strains and variability in a successive outbreak has been observed in Africa as previously reported by Safa, et al. (2008) with such variant strains exhibiting increased virulence probably due to their acquisition of extra characteristics. Table 1 show a detailed case fatality of the V. cholerae member since the past twenty-two years validating that, the need for a continuous surveillance study cannot be overemphasized (Tables 1-6) (Figures 1 & 2).
Table 2: Global Variability Trends in Vibrio Cholerae Occurrence.
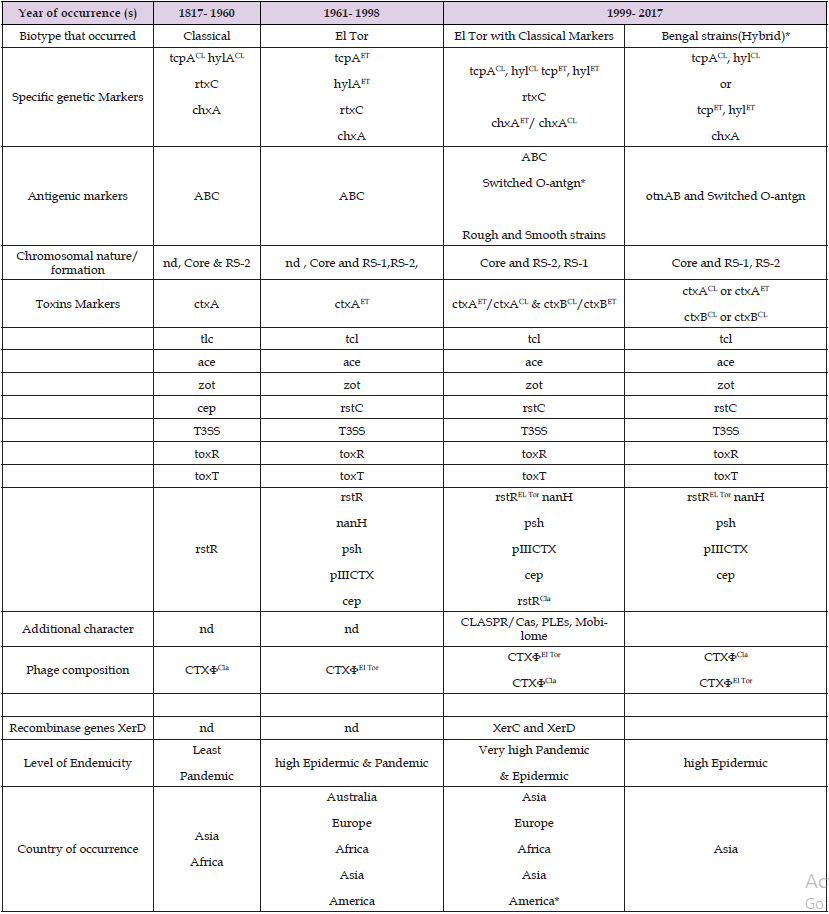
Note: *= It varies, nd= not determine
Table 4: Analysis variance of the effects of soil texture and water stress on the growth of potato fresh weight.
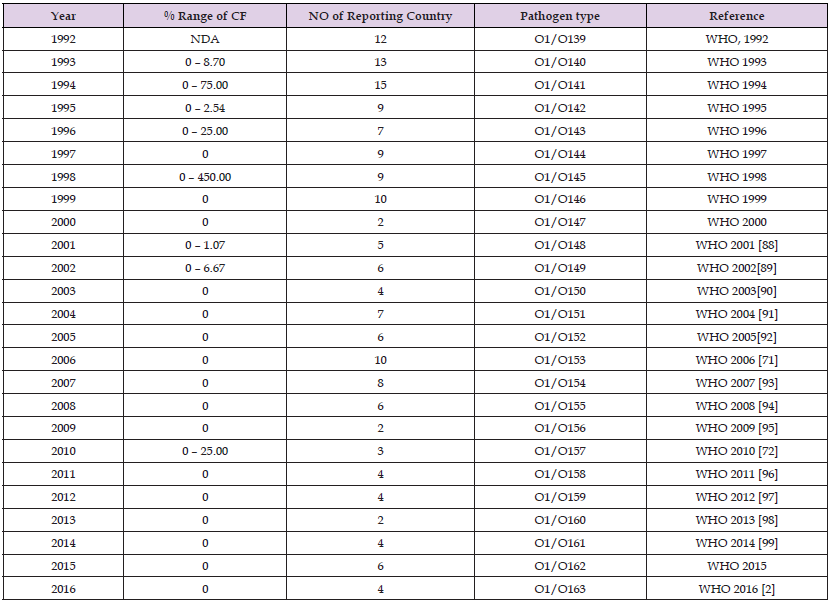
Case Fatality in Asia
In the Asia continent, an alarming epidemic of V. cholerae has been a continuous recurrence since the emergence of cholera epidemic and pandemics (7PETs). As reported, it was said to have started in the Ganges Delta region and spreading in a course of time to other continents, killing many affected persons (Pollitzer, 1954). It is noteworthy to emphasize that all previous six pandemic recorded originates from the Asian continent. The seventh pandemic and the one with the highest geographical spread/duration arise outside the Indian sub-continent in early 1961 in Indonesia (Island of Sulawesi) spreading to neighbouring countries with a causal agent later characterised as V. cholerae O1 biotype EI Tor (Cvjetanovic and Barua, 1972; Plaza, et al. 2018). The study of Pollitzer (1959) pointed that the causal agent was first observed and isolated in an Egyptian village called EI Tor in 1905 before its later occurrence in Sulawesi, Indonesia in 1937 (Pollitzer, 1959; Tanamal, 1959). It continued to spread amongst developed and developing countries in the world today eg Bangladesh, Rwanda and Mozambique (Sack, et al. 2004).
Newer strains of cholera epidemic were reported between 1960 and 2010 resulting from V. cholerae O139 sero-group from Calcutta, Bangladesh and other areas. It is suspected that as a result of the changing nature of the pathogen, there is possibility that it would herald the eighth pandemic (Didelot, et al. 2015). This cholera pathogen was later replaced by the previously reported O1 sero-group with different genetic characteristics (Mukhopadhyay et al. 1996; Faruque, et al. 1997; Yamasaki et al. [78]; Ali, et al., 2015). In late 1996 an outbreak showed a resurgence of a genetically diverse and an anitibiogram altered strain of O139 sero-group which dominated the O1 sero-group in Indian subcontinent. This strain was said to possess different characteristics from all O1 and O139 that were previously reported in 1992 (Yamasaki, et al. [78]). In another study, reported data showed that another member dubbed Bengal O139 sero-group, arose from sero-group O1 biotype EI Tor and acquired genes from non-choleragenic strains (Mooi and Bik, [35]). Other related non-O1 strain observed shows pre-immunity selective advantage suggesting their variant tendency in adverse condition. Since then, there has been several prevalence report of V. cholerae variants in the Asian continent (Taneja, et al. 2009; Okada, et al. 2010), which may be probable pathogens of the recent pandemic reports (7PETs). The pathogen (7PETs) has been described to be implicated in a range of 0 – 11.7 case fatality in 2015 and 0 – 1.9 percent case fatality in 2016 at all 12 Asian countries (WHO [1,2,79]).
Case Fatality in Australia
There has not been much documented report of reoccurring outbreaks in Australia. Most sparing reports shows that it resulted one death which may have originated from travelled carrier in contaminated food (Didelot, et al. 2015). Recent occurrence of V. cholerae in Australia reported 2 cases and with no recorded deaths (WHO [1]). The study of various investigators showed that V. cholera members are found in areas of Australia where Vibrio are not endemic. The pathogens occurrence is mainly associated with environmental influence than anthropogenic activities (Giovana, et al. [80-85]).
Case Fatality in Europe
Between 1992 till date, Europe has not recorded much of V. cholerae epidemics, although the few that occurred recorded the highest known percentage case fatality in 1998. Some other years with high percentage case fatality are 1994, 1996 and 2010. The identified causal strain was the EI Tor O1biotype. The most recent recorded event had the strain having new acquired pathogenic and resistant genes which is different from genes present in previous recorded reports (WHO [2]). From 2003 till today there had been sparing occurrence V. cholerae as only six countries in Europe reported imported case in 2015 (France, Norway, Sweden, Spain, United Kingdom and Switzerland) (Rashed, et al. 2013; WHO [2]).
Case Fatality in America
Vibrio cholerae disease was first reported in the less developed region of Latin America where there was no previous pandemic report in 1991. It was reported that the seventh pandemic did not affect these region, but the epidemic occurred along the Pacific coast of Peru spreading into the Southern and Central countries of American continent such as Ecuador, Colombia, Brazil, Chile and central Mexico. According to the Pan American Health Organisaton, over 750,00 cases was reported with 0.87%(6500) death recorded between 1991-1992 (Tauxe, et al. [86]; WHO [2]). Although it was reported by Karaolis et al. [87] that the first occurrence of V. cholerae was in the United States Gulf Coast in 1991, a coincidental El Nino of tropical Pacific related event occurred where the isolated organism was a non-choleragenic V. cholerae O1. The pathogen showed characteristic difference from the endemic strain in USA with several similar characters as the causal agent of the seventh pandemic (7PETs) (Domman, et al. 2017; Didelot, et al. 2015). A multilocus enzyme electrophoresis comparing the strains further confirms the distinct clone and its similar region suggesting their relatedness.
Hence, it was said to have originated from choleragenic strains which through the mechanism of horizontal gene transfer lost the known and detectable virulence factors (Plaza, et al. 2018; Fri, et al. [27]). It was also shown to express other newer pathogenic factor necessitating their form as an emerging strain (Clemens, et al. 2017; Ali, et al. 2015). This observation has been previously reported after the study of Bik, et al. [35] Bik and his group reported an emergence of O139 sero-group that originated from O1 sero-group after a horizontal gene transfer to a non-O1 sero-group previously observed. From the ongoing reports, it is expedient to study the pathogens variant nature for a better knowledge and awareness. The study is also necessary to explain the integrity of reoccurring newer strains and future preparedness of intervention strategy to manage any epidemic outbreak as illustrated by the Peruvian epidemic report. Shown in Figure 1 below is a map of various affected regions by the seventh cholera pandemic and its spreading direction onto various regions in the world between 1960 and 1991. According to WHO [1], only two major outbreaks of cholera have been note in America since the last fifty years which were associated with travellers migration. The United Nation peacekeeping team were not left out in the spread as the strain isolated in the recent outbreak shows a striking association with the Asisn strain which may have been carried to America by migration [88-99].
Historical evidence shows that dynamism amongst successive Vibrio population is a fact as such changes have been reported from the various pandemic from first to seventh (Kaper, et al. [5]; Stine, et al., 2014). Similarity in ribotype was reported in a Vibrio member classified as classical biotype from the second pandemic and a clinical specimen although there were not able to strain type the first pandemic Vibrios due to few available literatures and advance typing systems. The last two pandemic were show to be caused by both classical and EI Tor strains (Faruque and Mekalanos, [100]). One observed recurring event in any outbreak is the disappearance of a previously prevalent strain from a previous outbreak and an emergence of newer strain with severe pathogenesis as depicted in Table 1 (Mutreja, et al. [5,56,63]).
Serological determination of smooth and rough variants of V. cholerae is dependent on the nature and repeated lipopolysaccharide (LPS) units (O-antigen). Various epidemiological roles of the LPS and O-antigen, including its protection and adhension dynamics have been reported by various investigator (Wachsmuth and Olsvik, 1994; Kaper, et al. [6]; Islam, et al. 1994; Gu et al. 2014). The serologic scheme emphasize on the availability of paired allele of smooth (S) and/or rough (R) ‘O’antigenic determinant. Vibrio strains that are tagged rough (R) are those members that possess in their LPS similar rough ‘O’ antigen paired allele. Those that possess two dissimilar smooth and rough O-antigen allele are tagged smooth strains (Sakazaki, 1992; Keya, et al. 2004). A study on rough mutant smooth strains of V. cholerae shows a severe bowel defect with reduced fluid accumulation (Angelichio, et al. 1999; Mitra, et al. 2001; Keya, et al. 2004). The expression results from a core-A attachment of the O-antigen to the LPS which determines smoothness but the absence of such core-A attachment results a rough.
However, there has been report of some rough strains whose character does not depend on the presence of ‘O’ antigenic determinants. There has also been report of smooth strains conversion to rough strains resulting change in the morphological characteristics of the affected strain, traced to harsh environmental conditions (Rochetta, et al. 1999; Islam, et al. [101]; Igbinosa, [19]; Hasan, et al. 2012). Increase and decreased pathogenesis of the rough strains has also been reported by Islam and collegues (Islam, et al. [101]). Their result presents diarrhoea induction and a reduction in diarrhoea fluid accumulation/production in rabbit ligated ilea loop test (RLILT) for the rough strain when compared to fluid accumulation from similar RLILT in a smooth strain. This report is an evidence of exotoxin production and the cell wall LPS different arrangement of the rough strains when environmental conditions become favourable. Previous studies of Wachsmuth and Olsvik (1994), also show similar report, a possibility for reversibility of rough to smooth strains and vise vassal, which is associated with adaptive response. In another study by Makin and Beveridge (1996), it was reported that the B-band O-antigen was lost from Pseudomonas aeruginosa strain (PAO1) LPS when its cells were incubated at reduced nutrient, higher temperature (45 oC), high salinity (NaCl, MgCl2), low pH and glycerol. This indicates a lose in the O-antigen synthesis at such unfavourable conditions and an appreciable increase in the A-band O antigen (McGroarty and Rivera, 1990; Makin and Beveridge, 1996). Following these reports, the need to emphasize research on such environmental factors and adaptive response on rough-smooth conversion of strains as well as variability is imminent.
Other relevant contributors to the variable nature of V. cholerae across regional and historically important strain is the report of the comparative whole-genome study as it provided important evolutionary details of the pathogen (Chun, et al. [5,102]; Turner, et al. 2017). Phage’s are said to be genetic elements which infect bacterial by possibly searching specific host genomic DNA and enter by insertion to form bacteriophage. Its life cycle revolve around dimorphism in the form of a temperate phage or virulent phage. The temperate phage integrate its genetic material into the host genomic DNA to become part of the host total genomic make up and express lytic and lysogenic functions (Prophage) (Shlezinger, et al.; Yen et al.2017; Igere and Adeola [103]). Whereas the virulent phage express a lytic character which results the release of progeny phages hence it contribute to the pathogenicity of infected hosts through a lysogenic conversion (Faruque, et al. 1997; Brussow, et al. 2004; Leitet, et al. 2006; Gorski, et al. 2017). The evidence of phage, its carriage of cholera toxin genes and related regulatory appendages that mediates phage conversion of non-toxigenic strains to toxigenic strains has also been discussed (Waldor and Mekalanos [5,28,39]; Yu et al., 2013).
Apart from the point mutation in ctxB gene exhibited in CTX phages, one observed marker is the replacement of phage carrier by the pathogen and reproduction of variant CTX phages such as CTXcla or CTX-1 and RS1 which is a satellite phage (Sarkar, et al. 2011; Raheed, et al. 2013; Rajpara, et al. 2013). These are some of the uncontrolled genetic activities which regularly occur, resulting Vibrio population variability. The presence of lysogenic phage in Vibrio members has been associated with the variability of these strains as reported by Faruque, et al. [11] and Kim, et al. (2015). Due to the survival dynamics initiated by the pathogen in aquatic environment, a process of adaptation occurs at high pH condition, increased salinity, temperature above optimal range, nutritional depravity and presence of bacteriophages resulting the activation as well as expression of numerous/ diverse genes (Makin and Beveridge, 1996; Poirier, 2012; Shah, et al. 2012). In addition to these, a rugose state or viable but non-culturable state is attained by the pathogen.
Phages or bacteriophages are dubbed viral-like bacterial which consist of mobile genetic elements. Such elements are involved in horizontal sharing or genetic transfer which are major contributors to evolution and re-emergence of Vibrio strains (Poirier, 2012; Bompangue, et al. 2012). It was also reported in another study that phages uses the toxin co-regulator pilus (TCP) which is associated with the virulence genes as an anchor onto non-choleragenic strains making them to become pathogenic during a lysogenic conversion process (Igbinosa et al., 2009; Plaza et al., 2018). Other report shows that the phage genome harbours some novel virulence genes that may promote possible re-emergence (Plaza, et al. 2018; Sarkar, et al. 2011; Naser, et al. 2017). A notable example of such toxin – harbouring - phage is the CTXØ (Waldor and Mekalanos, 1996; Naser, et al., 2017). It possesses a variant form of the ctx gene (ctxAB) which ensures patho-conversion of Vibrio strains that acquire it irrespective of its previous nature (Alejandro, et al. 2016; Faruque and Mekalanos, [104]). Phage’s were first observed by d’Herelle in 1926 in bacterial strains but much attention was not given to its study until 1950 when several distinct clones of phage’s were described amongst Vibrio cholerae strains (Ackemann, et al. 1984; Faruque, [105]).
The detection of phage as a typing system in microbial differentiation scheme and strain characterisation has to a great extent ensured the epidemiological study of the pathogen (Faruque [105]; Kim, et al. 2015). Since its application in surveillance study on the spread of the EI Tor related cholera outbreak, it has been used to classify various O1/O139 biotypes, although not without limitations (Mukerjee and Phil, [106]; Chakrabarti, et al. 2000; Sarkar, et al. 2011; Naser, et al. 2017). Lysogenic phage’s today have been tagged as contributors to the changing virulence pattern of O1/O139 Vibrio cholerae in successive emergence especially in cholera endemic regions (Bangladesh) as well as influencing seasonal/duration and severity in outbreaks (Faruque, et al. [12,107]; Faruque and Mekalanos, 2012). The aquatic milieu is not left out as it is a major precursor to the survival of non-O1/non-O139 Vibrio strains (Isaac-Marquez, et al. 1998; Faruque and Mekalanos [104]; Sarkar, et al. 2011; Dutta, et al.2013). Phage’s among non-choleragenic Vibrio has shown similar properties in inter-epidemic periods (Alejandro, et al. 2016; Elhadi, et al. 2012; Das and Gupta, 2005).
As a result of the numerous characters acquired by phages as reported from genomic studies, single nucleotide protein (SNP) and characterization of phages (Alegandro, et al. 2016), they are classed by the rstR and ctxB genotype (Das and Gupta, 2005). It was reported that CTX phage and RS1 were incorporate into the Vibrio chromosomes (chromosomal deconcatination) through a deletion-induce filametation controlled by dif sequence and a recombination site Xer (Das, et al. 2013; Elhadi, et al. 2012). The phage genome recombination results replication of phage CTX and toxin-genesis (Turner, et al., 2017). Although it has not been fully documented, phage CTX and RS1 share similar genes (rstREI Tor, rstA and rstB) (Davis and Waldor, [108]; Park, et al.,2011; Alegandro, et al. 2016). This is demonstrated in an excision studies on CTX-1 and RS1 from the vibrio chromosome 1 of the EI Tor strain using recA mechanism, which is said to be enhanced by rstC in both in vitro and in vivo activity (Kim, et al. [36,109]; Lin, et al. 2012). These reports shows the presence of the rstREI tor and rstRcla among various strains (Das, et al. 2013, Kim, et al. 2015), as the rstRCalcutta was characterized in the CTX gene of O139biotype while the rstRenv gene was identified from environmental V. cholerae strains (Nusrin, et al. [110]; Yu, et al.; Alegandro, et al. 2016). The toxin gene in the classical biotype (CTXcla) were shown to consist of rstRcla and ctxB1, while the remaining phage genome differs from the EI Tor strain CTX phages by possessing numerous SNPs (Chun, et al. 2006; Lal, et al. 2016; Katharios, et al. 2017).
The studies of Waldor, et al. [39] and Zhang, et al. (1995) have shown that one or more CT genes are encoded in Toxigenic V. cholerae dubbed ctxA and ctxB. These A and B subunits gene copies of CT are encoded by two different but overlapping open reading frames (ORFs) within the phage genome. V. cholerae also produces other putative toxins known as zonula occludens toxin (Zot), which ensures increase in the permeability of the small intestinal melius. It also influence the structural determinants of intercellular tight junction, called zonula occludens (Baudry, et al. [111]; Fasano, et al. 1991). Another described toxin is the accessory cholera enterotoxin (Ace) which possess the ability to induce excess fluid accumulation in rabbit ligated ileal loops test (RLILT) (Trucksis, et al. 1993). In toxigenic V. cholerae, CT is encoded by filamentous prophage or vibriophage designated CTXΦ, which exists in the bacterial chromosome. These reports were also resently confirmed by numerous experts in Vibrio studies (Fri, et al. [27]; Davies, et al. 2017; Naser, et al. 2017; Okada, et al. 2018; Kalatzis, et al. 2018). The existence of CTXΦ is unusual amongst filamentous prophage’s because the phage genome encodes the functional site specificity integrational system and thus can integrate into the V. cholerae chromosome at a specific attachment site known as attRS, forming stable lysogens (Waldor and Mekalanos, [39,101,104,112]; SarKar, et al. 2011; Yu, et al. 2013; ).It is important to note that in a standard typing system for detection of pathogens, phage typing cannot be omitted or avoided. Phage typing has been a routine practice since the application of microbiological methods in the detection of pathogens especially those of the enteric family which Vibrio members belong. Recently most investigators tend to bypass or omit the application of phage typing which portend substandard application.
One observed variability amongst V. cholerae detected in the various region of pandemic or epidemic was antibiotic resistance. Currently, their prevalence is on the increase especially amongst Gram-negative bacteria, upon which clinical management/control (treatment) options has failed (Kuma, et al. 2014; Raissy, et al. 2012; Igere, et al. [113,114]). Particular and predominant amongst them are the ones expressing protease resistant genes coding for New Delhi metallo-β-lactamase 1 (NDM–1). Others are the Cephamycinase, Extended Spectrum beta lactamase, oxacillinase as well as members of other serine and metalo-β-lactamase (Rao, 2015). Within the last two decades, such resistance have been upgraded onto pan drug resistance (PDR) and extensive drug resistance (XDR) as recently reported by infectious disease society of America (IDSA, 2012). Research attempt towards ensuring possible reversal of such resistance revealed low outcome (Jacqueline [115]; EMA, 2012; Sundqvist, et al. 2010). Early studies have documented an association of antibiotic resistance to both pandemic and epidemic cholera outbreaks.
The study of Glass, et al. [45] revealed an emerging strain with variability amongst transient multiple-drug-resistant (MDR) V. cholerae. Strains of O1/O139 have been reported to have produced varying antibiogram (Siddique, et al. [116]; Kitaoka, et al. 2011). The study of Waldor, et al. [39] on O139 serotype showed acquired resistant genes to the common control antibiotics (Sulfamethoxazole-Trimethoprim or SXT and Streptomycin) for which it was previously susceptible in the 1992 outbreak in Bangladesh. By 2002 and other preceeding years, Vibrio members re-emergence reported multiple antibiotic resistant to tetracycline, streptomycin, sulfamethoxazole-Trimethoprim and erythromycin with O1 strain reported as predominant (Raissy, et al. 2012; Kitaoka, et al. 2011; Kuma, et al. 2014). As a follow-up to previous outbreaks and the reported resistant nature, a study conducted by Mitra, et al. [13] showed a similar occurrence in their antibiogram when strains from 1992 out breaks were compared with strains of 1997. In a similar study on the genetic variability of the SXT and streptomycin resistance, it showed an approximate 3.6-kb deletion of the integrative element among the susceptible strains while a deleted region was not found among the resistant strain depicting an insertion and gain of function mutation (Kitaoka, et al. 2011; Baron. et al. 2016; Dengo-Baloi, et al. 2017). The SXT or Sul-1 element is seen to be associated with self integrative element which is spreading amongst Gram negative especially the O1 Vibrio members and also implicated in a Bangladesh epidemic outbreak (Faruque, et al. [11]; 2005; Baron et al., 2016). It is suggestive that occurrence of such mutation is the origin of multiple drug resistance amongst Gram negative organisms.
An observed change in the antibacterial resistant pattern or the antibiogram was previously reported by recent investigators which indicate a substantial mobility of genetic elements in O1 strain of V. cholerae (Faruque, et al. [7,23]). It was observed in the study of Rashed, et al. (2013) that some members of ctxBET strain of O1 V. cholerae are closely related to previously reported 2001 strains. In similar manner, there was similarity in the ctxBET of O1 and O139 V. cholerae strains compared to those in previous studies of 1993. The similarity spans into their antibiotic resistant band patterns and their restriction enzyme digest band pattern when NorI endonuclease was used. However, it is worthy of note that the susceptibility of O1/O139 choleragenic Vibrios is higher with the fluoroquinolone antibiotic members (Yamamoto, et al. [117]; Dengo-Baloi, et al. 2017). Hence those members of antibiotics are useful in the treatment of Vibrio implicated cases, although there are possible tendency for development of resistance to these antibiotics. Interest in unravelling these antimicrobial determinants and multiple drug resistant traits expressed by these bacterial shows the presence of a possible transfer of a horizontal conjugative transposon (antibiotic resistant integron) into V. cholera. This explains the reason for expressing such antibiotic resistant markers as it utilizes the antibiotics. It also explains the variability in antimicrobial determinants as well as the instability and diverse state of O139 sero-group genome, hence the fluctuation of their next possible outbreak or emergence in any area.
Acquisition of plasmid (extra chromosomal DNA) amongst multiple antibiotic-resistance strains of O139 V. cholerae that belong to incompatibility group C has been reported (Yamasaki, et al. [78]). In the study of Mandal, et al. [9], it was reported that the O1 V.cholerae of the Ogawa serotype which was associated with the cholera outbreak in Kolkata, India harboured multiple drug resistant plasmid. The emergence of plasmid mediated antibiotic resistance has been shown to be associated with the changing pattern of antibiogram. Several investigators are insinuating plasmid involvement in the variability or diverse nature of V. cholerae strains (Amaro, et al. 1988; Colombo, et al. 1997; Misra, et al, 1998; Thungapathra, et al. 2002; Zhang, et al. 2012; Poirel, et al. [118]; Letchumanan, et al. 2015).
Earlier in 1986, Hamood and his colleagues reported the the presence of plasmid in a Vibrio strain results to changes in antibiogram pattern and virulence (Hamood, et al. [119]). In another similar study by Carraro, et al. [120], it was reported that IncA/C conjugative plasmids which was present in the integrative and conjugation elements (ICE) (among the SXT/R391Mobilome family), act as agents that enhance integration of novel genomic island (GI) in V. cholerae strains (Mala, et al. 2017). Other mobile genetic elements that encodes prophage’s and cholera toxin in the vibrio pathogenicity island are also associated with the plasmid mediation. The presence of the IncA/C conjugative plasmid was also reported to be associated with multiple drug resistance amongst V. cholerae as well as the variability observed in successive clone from previous outbreaks (Carraro, et al. [32,120,121]).
Abiding with the various reports on the diverse nature of V. cholerae in successive pandemic, one may be quick to say that studies on the genetic mechanism governing the change is a key to understanding such occurrence. However the exact situation remains vague as its driving force is yet unravelled. The immunity of previously infected persons and environmental factors in endemic regions, are presumed to pose possible factors as reported by various investigators (Karlsson, et al. 2013; Yoon and Mekalanos, [122,123]). Pathogens interaction with human, environmental influence and genetic mechanism/ effect of the pathogen are presumed as source to the generation of variant strains. However, genetic influence alone does not explain ‘the how and why’ of occurrence.
Hence it is better explained by a consortium of determinative strategy which may include the
(i) Immunity factor of host, which discuss the management/control strategy employed in the previous outbreak and how host responded to such treatment,
(ii) Factors of the environment that herald variant formation. This explains the activity of the pathogen in the aquatic environment as exemplified in the changes that occur between 1961 and 1990 where the classical strains in the ocean change to the EI Tor strains. Suffix to say that the sixth pandemic recorded the classical biotype strains for both clinical and environmental specimen and
(iii) Genetic mechanism of pathogen. It deals with the lysogenic phage conversion, plasmid profile, and genome dependent toxin regulatory appendages as well as the CTX dynamics. It was also observed from the study of Bakhshi, et al. [124] that some unidentified variants may have occurred in the Ocean as depicted in the results from the Iran 2012 outbreak and Bangladesh outbreak between 2011-2012 in areas were the EI Tor strain was not previously recorded among clinical specimen since 10 years prior the study (Rashed, et al. [124,125]). These results shows that present in the environment are also strains-in-wait for possible driving force to change. Another very important factor that may influence and is presumed to influence variability amongst Vibrio species is the availability of mobile and cell free DNA (mcfDNA) in the aquatic environment (Zhang, et al. 2017). According to investigators, such cell free DNA are basically the driving force of gene transfer and multiple antibiotic resistance mechanism, as such it has been a neglected area of study especially in the study of Vibrio.
Microbiological detection of O1/O139 and non-O1/non-O139 V. cholerae is conventionally culture-based, were suspected samples are cultivated on selective solid media after an enrichment procedure. Such selective media are thiosulphate citrate bile salt agar (TCBS) and/or Monsur taurocholate tellurite gelatin (MTTG) agar while the enrichment media is alkaline peptone water (APW) (Hasan, et al. 2012). Growth yielded colonies of presumptive isolates after 18 - 24hours incubation are thereafter observed for their morphological (Grams reaction) and cultural characteristics (motility and forms), which will be followed by a battery of biochemical reactions, sugar fermentation, serology, stability with chemical agents as well as thermal stability and invasiveness of expressed virulence factors using rabbit ligated ilea loop test and guinea pig eye test (Huq, et al. [15,19,126]; Rao, 2010; Begum, et al. 2006).
Recently, advanced methods now employ Matrix Assisted Laser Disorption Ionisation Time of Flight Mass Spectrometry (MALDI-TOFMS), PCR detection or characterisation and other molecular biology techniques such as amplified fragment length polymorphism (AFLP) or restricted fragment length polymorphism (RFLP). Other detection test may include cellobiose polymyxin sensitivity, blood haemolysis and chemical reactivity with Sodium deoxycholate, cholera red test and ribotyping (Arias, et al. [127-130]). The application of molecular biology techniques has reported high sensitivity and specificity in detection or identification of bacterial strains. One essential precept of such application is the use of 16SrRNA gene detection for identification amongst bacterium (Priyanka, et al. [130]). It is an appropriate routine choice technique to study both the inter-genic and intra-genic relationships as well as variable region of bacterium such as Vibrio members (Kim, et al. [131]). Applying molecular biology techniques also provides results of epidemiological and phylogenetic relevance, comparing and assaying the differences in variant organism among choleragenic and non-choleragenic vibrio cholera (Espineira, et al. 2017; Saidi, et al. [132]).
In addition to the above molecular biology techniques, it is suggestive that methods of detection with increased microbiological relevance such as antibiotic resistant pattern, plasmid profiling, phage typing and ribotyping be applied amongst investigator and practitioners. These methods of detection are applicable to emerging, re-emerging and recalcitrant pathogenic organism of increase clinical relevance with high case fatality in both reference and quality control laboratory (Igbinosa, et al. [13,19,130]). It is noteworthy that laboratories with expertise in all the above listed detection methods, should apply all techniques routinely to ensure that such epidemic outbreak related pathogens are detected and controlled with proficiency to avoid recalcitrance. Their stringent application will enhance understanding of the variability and track the origin of variability. It will also reveal the changing nature of such pathogens at every successive outbreak case in our environment as reported amongst the O1/O139 and non-O1/O139 strains/variant of choleragenic V. cholerae. The trend of such changing activity may also be envisioned or predicted which will also enhance control of re-occurence.
The Vibrio cholerae members today continue to evolve with variant strains possessing newer pathogenic indices. Previously it was reported that a small chromosomal DNA has been attach to the Vibrio genomic Island which ensures its mediation for variability and higher pathogenic potential in addition to the insertion of prophage region (Faruque, et al. 2000; Kim, et al. 2015). Current reports have shown that apart from multiple antibiotic resistant natures of the members, the pathogen has now acquired mobile genetic elements, advanced form of phage and plasmids (Bellanger, et al. [23,112,133-135]) reported that Vibrio and other Gram negative/positive organisms now possess a self defence mechanism against mobile genetic dynamics and phage integration. A mechanism exhibited by the classical biotype members, is encoded by the Clusters of Regularly Interspaced Short Palindromic Repeat (CRISPR) and other CRISPR associated proteins (Cas). It is said that the genomic DNA of over 40% bacterial and 90% Archea now possess the CRISPR/Cas as interpreted from the whole genome sequence studies (Seed, et al. [11,136]; Box, et al.).
Suffix to say that the CRISPR/Cas loci is use by Vibrio member to dislodge invading agents such as DNA/RNA, viruses, chemical agents (Bellanger, et al. 2013; Marraini, 2015). Further studies by various investigators shows that in a bid to survive the effect of the CRISPR/Cas activity, the phage acquires a loci into its genome to compromise the effect of the CRISPR/Cas structure and continue to thrive/replicate with the organism as reported in V. cholerae (Dedrick, et al. [136,137]; Box, et al., 2016; Marraini, 2015). The acquisition of newer character by the vibriophage over time necessitates vibriocidal potential and its usage in control of cholera. The struggle for supremacy continues as V. cholerae now utilises the replication of phage to elicit a novel chromosomal island that behaves as the phage which is called phage-inducible chromosomal island (PICI) (Seed, et al. [112]). It occurs when a vibriophage trys to infect a V. cholerae, hence the name PICI or PICI- like element (PLE) (Bondy-Denomy, [112,138]).
When PLEs in V. cholerae try to chunks the replication of phage, the phage which has previously acquired CRISPR/Cas structure then fights back and establishes its survival in the pathogen (Bondy-Denomy, [139-141]). These effect are not limited to phage acquisition of genetic structures only, the effect of other mobile genetic elements is also another variable state of V. cholerae today. There are anticipation that in the near future, some genetic character may be acquired by the V. cholerae to compromise the activity of variant phages, else the variability of next successive strains may be disastrous. The future remains unpredictable as the pathogen continues evolving; the need for advance/improved detection scheme which will enhance management and control cannot be overemphasized.
Most importantly, early diagnosis and clinical management are observed gabs in the control of cholera outbreak cases. Research on vibrionaceae, though has some limitations, has been able to provide solution to immediate control mechanism for cholera epidemic. Availability of relevant diagnostic tools as well as identification scheme has been lagging due to cost effectivity, technical expertises of such techniques and the equivalent advanced research in vibrio studies as the pathogen evolve. Although diagnosis in other related cases seem to have advanced application, the cost of material remain high (Brittain- Long, et al. 2011; Endimiani, et al. 2011; Levy, et al. 2012). Yet, the diagnosis and detection of other pathogens/dieases such as detection of Streptococcus pyogenes are available (Burkhardt, et al. [142,143]). Cost reducing strategies have not yielded the expected results. Early detection of cholera is one possible aspect that has not been extensively analysed. It is assumed that point –of-care (first point of suspected case) diagnostic tools would ensure early detection of cholera suspected cases especially in cholera endemic low-and-middle-income- countries (CELMIC).
One of the aspect that has not received much attention in vibriology is the global comperative genomic research. Expected authorities that may have elucidated the variability of Vibrio members as reported by various experts seem to be negligent or had a probable reduced interest in such research. The application and distribution of Vibrio vaccine into endemic regions would have been controlled by the understanding of any countries history of out break, emergence and re-emergence. The interval (time/period) within succesive outbreaks as well as the trend of variability may also be predictable. It is suggestive that guided by this understanding, the distribution of the vaccine and application may be efficient. Another aspect that has not received much attention is clinical management. It is observed today that the pathogen (V. cholerae) is evolving regularly and spontaneously necessitating interest for rapid diagnosis and stringent management. Reports from various investigators show that multiple antibiotic resistant amongst the vibrio variant is on the increase (Okeke et al. [144]).
Strategic improve rapid diagnostic methodology must be designed, implemented and validation of new diagnostic test be employed to meet the need of point-of-care diagnosis. The application of which will surely control the spread of cholera infections when diagnosed early. The challenge of cost should also be taken to note while implementing such improved methods. It is suggested that such method of detection must include but not limited to Phage typing, antibiotic resistant profiling and plasmid profiling (Nordmann, et al. 2012). Effects towards accessing the most needed antibiotics must also be employed (Norrby, et al. [145]). The application of residual chlorine in wastewater treatment and the respective increase in the amount of chlorine in treated water had been traced to the rugose existence and viable but non-culturable nature (VBNCN) of the organism (Beyhan and Yildiz, [146]). Local/global support partnership must be strictly adhered to urgently and specifically, as suggested by the WHO/ Global Task Force on Cholera Control (GTFCC) (WHO/GTFCC, 2017).
The future remains vague since variability observed amongst successive V. cholera clones in outbreak continues to threaten control and treatment/management strategies taken by the public health systems. The variability paradigm has been show to be spreading beyond research advancement over time. The surge of V. cholerae O1/O139 and Non-O1/Non-O139 serogroup has not been adequately ruled out of recent pandemic outbreaks as it has been implicated in over 30,000 infected persons in Asian and African continent. Most investigators have reported over 26 variants with specific identifying gene for each member. Current debate is emphasizing/suggesting that these pathogens may herald the eighth pandemic of cholera if not controlled. As a global roadmap to elimination of cholera, an early detection and multiple sector surveillance screening must by applied. The need for urgent inclusion of a complete classification scheme is on the cards.
This scheme should include all known microbiological methods and mobilome typing, phage typing, plasmid profiling, genomic island characterisation, phage genomic characterisation and whole genome sequencing of any pathogen that present a suspected case earlier enough in rural cholera endemic areas. The application of the aforementioned with standard molecular biology and biotechnological techniques will ensure the specificity of results. The spread of these variant re-emerging strains of V. cholerae should be carefully monitored employing Strategic environmental and epidemiological surveillance Scheme (SEESS) as properly designed national organismal characterisation scheme remains the best approach. The urgency, enforcement of these detailed standard detection techniques and early report of cases must not be treated with levity [147-203].
The application of whole cell attenuated vaccination, which is a basic control strategy, may in the future fail in some outbreak of cholera endemic region within the globe due to the changing nature of the pathogen with various variability factors (change in antigenic determinant). A continent based molecular and/or genomic characterisation (comparative genomics) as well as continent based vaccine development and application may pose a relevant control strategy. There is also a suggetive need to ensure the discouragement of any inter-continent transfer of such vaccine. Such coordinated partnership dependent approach (consortium) will involve various countries and continents politics, mobilization of social, policy makers, synergy of locals wellbeing and experts technical relevance. The application of these schemes may be principal to the control of re-emerging variant of V. cholerae.
The occurrences precluding Peruvian epidemics as reported by Anon, in 1991 had also necessitated the needed preparedness for possible future outbreaks. Finally, studies are ongoing to analyse both clinical and environmental strains from different areas of South Africa to analyse the various strains in these regions with a view to unravelling the nature of their respective genomic island, Phage type and genomic characteristic, mobilome, PLEs, drug resistance, CT domain, ToxR-ToxT and TCP gene cluster, their structural dependency and variation in their pathogenicity.
Although reports from various investigators in the world have shown that the major progenitor of V. cholerae variability has been described as vibriophage. It has been recognised as a human ally that can/may help in the control of possible cholera outbreaks. However, the case fatality remains very high especially amongst developing countries, hence applying a well studied vibriophage promises future therapy and better control mechanism. Other recently identified character of V. cholerae such as CRISPR/Cas and PLEs and the acquisition of these genetic elements by vibriophage promises a future for the control of cholera. Incorporating the variant vibriophage into drug regimen may serve as a defense against cholera outbreaks. This ingenious vibriophage attributes could be harnessed through endless strategies including probiotic products, edible-fruit antibiotics, alternative treatment both in human health, animal and agriculture.
The emergence and re-emergence of newer strain and its pathogenicity still remain a topic of public health debate. Few genomic sequence data are available from Vibrio studies. A possible starting point for future research may be based on examining a continent base comparative genomic sequencing and phylogenetic studies of V. cholerae. A developed surveillance scheme which will include phage typing, mobilome typing, characterization of lysogenic and temperate phage’s, plasmid profiling must be strictly upheld. Detection of polymorphism amongst the CTX and other associated toxigenic genes within the Vibrio pathogenicity Islands (VPS-1, VPS-2) and genomic island (GI) must be adequately assessed. Suffix to say that the observation of phage implication in relative genetic conversion and infection of cells as previously reported by other investigators/ researchers is not left out. This would serve as a possible stand-point for future research with an aim of understanding the mechanism of such gene transfer, sero-conversion, pathogenesis and polymorphism amongst related pathogenic genes. These analyses are currently ongoing within our laboratory investigations.
The need to focus on stringent identification/characterisation scheme which is based on application of molecular biology techniques and a possible surveillance strategy for successful control is eminent. Application of such standard microbiological methods with improved surveillance scheme and early report of suspected cases pose promise for a future control of cholera outbreak. Results from such studies may provide solution to control or better knowledge on the variability and antimicrobial determinants observed amongst V. cholerae O1/O139 and non-O1/non-O139 members.
We are grateful to the South African Medical Research Council (SAMRC) and Govan Mbeki Research and Development Centre (GMRDC) of Fort Hare University for their financial support. We are greatful to Prof Uchechukwu U. Nwodo and Dr Ben Iweriebor C for the time to read through the manuscript, advise, attention and design given during the writing of this review.
Authors declared no competing interest.
The author(s) acknowledge the funds provided by African German Network of Excellence in Science 2022 (AGNES-2022) and Govan Mbeki Research and development centre.
The datasets/information used for this study are available on relevant request.
IBE: Conceptualisation, methodology, and investigation, data curation, analysis, interpretation writing-original draft preparation, writing-review and editing.


