Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Bodicherla Siva Sankar1*, D Natarajasivan2 and M Purushotham Reddy3
Received: August 02, 2024; Published: August 20, 2024
*Corresponding author: Bodicherla Siva Sankar, Research Scholar, Department of Computer Science and Engineering, Faculty of Engineering and Technology, Annamalai University, India, Email : s.bodicherla.sankar@gmail.com
DOI: 10.26717/BJSTR.2024.58.009131
Lung cancer originates from abnormal growth in lung cells, characterized by uncontrolled cell division within lung tissues. Early detection of lung cancer is crucial for improving patient outcomes and survival rates. The cited papers highlight several limitations, such as small sample sizes, reliance on previous studies, the need for future validation, resource-intensive methods, potential biases, lack of longitudinal data, necessity for further experimentation, limited clinical practice integration, dependence on imaging quality, and insufficient data for robust model training and evaluation. To address these limitations, suggested strategies include using CNN and RNN methods to compile larger datasets, confirming results through prospective validation, developing resource-efficient statistical methods, reducing biases through careful study design, conducting extended outcome evaluations, validating automated pulmonary nodule management systems, integrating multiscale features into breast cancer risk assessments, addressing imaging quality issues in healthcare, evaluating deep learning models longitudinally for EEG motor imagery, and using larger datasets to improve automatic lung nodule detection systems. With advances in machine learning, CNNs and RNNs have become powerful tools for analyzing medical images. This study aims to enhance lung cancer detection by comparing CNN and RNN models on X-ray image datasets. CNNs are known for their ability to identify complex image features, making them suitable for tasks like object recognition and segmentation.
In contrast, RNNs excel at processing sequential data, offering potential benefits in identifying temporal patterns in medical datasets. The study involves training and evaluating both CNN and RNN models on a dataset of X-ray images from individuals with and without lung cancer. We will assess each model’s performance in terms of accuracy, sensitivity, specificity, and computational efficiency. Additionally, we will explore the interpretability of these models to identify the features driving their classifications. Through this comparative analysis, we aim to provide insights into the strengths and weaknesses of CNNs and RNNs in lung cancer detection. Ultimately, our findings aim to guide the development of more effective and efficient diagnostic methods for early lung cancer detection, improving patient outcomes and reducing mortality rates.
Keywords: Lung Cancer Detection; Machine Learning; Convolutional Neural Networks (CNNs); Recurrent Neural Networks (RNNs); X-ray Image Analysis; Comparative Analysis
Lung cancer, marked by uncontrolled cell growth in lung tissue, poses a significant public health threat, emphasizing the importance of timely detection for better prognosis and patient outcomes. However, existing methods encounter challenges such as limited sample sizes, reliance on past research, the need for validation, resource-intensive techniques, potential biases, and insufficient data for robust model training [1]. To address these issues, leveraging advanced machine learning techniques like convolutional neural networks (CNNs) and recurrent neural networks (RNNs) holds promise. CNNs excel at analyzing complex image features, making them suitable for tasks such as object recognition, while RNNs specialize in processing sequential data, offering advantages in detecting temporal patterns in medical datasets [2]. Our study aims to enhance lung cancer detection by comparing CNN and RNN models using X-ray image datasets. We will train and evaluate both models on X-ray images from individuals with and without lung cancer, assessing their accuracy, sensitivity, specificity, and computational efficiency [3]. Additionally, we will explore their interpretability to understand the features guiding their classification decisions. Through this comparative analysis, we seek to shed light on the strengths and weaknesses of CNNs and RNNs in lung cancer detection, with the ultimate goal of improving diagnostic methods and potentially reducing lung cancer mortality rates [1-26].
The research methodology comprises multiple essential steps aimed at rigorously assessing the performance and interpretability of convolutional neural networks (CNNs) and recurrent neural networks (RNNs) in the realm of lung cancer detection using X-ray image datasets [4]. Initially, we will compile an extensive dataset containing X-ray images sourced from individuals both diagnosed with and without lung cancer, meticulously annotating each image to ensure precise and consistent labeling. This dataset will serve as the cornerstone for training and evaluating our CNN and RNN models [1-26]. Following dataset curation, we will preprocess the X-ray images to refine their quality and standardize their format, ensuring uniformity across the dataset. Preprocessing techniques, including resizing, normalization, and noise reduction, will be applied to optimize the input data for subsequent model training tasks [5]. Subsequently, we will develop tailored CNN and RNN architectures specifically designed for lung cancer detection. The CNN models will specialize in extracting intricate spatial features from the X-ray images, while the RNN models will focus on capturing temporal patterns inherent in sequential data [6]. These models will undergo fine-tuning using appropriate optimization algorithms and hyperparameter tuning methodologies to enhance their performance.
Once trained, we will conduct a comprehensive evaluation of the CNN and RNN models, employing metrics such as accuracy, sensitivity, specificity, and computational efficiency. Additionally, we will explore the interpretability of these models, utilizing techniques such as feature visualization and saliency mapping to elucidate the factors influencing their classification decisions [7]. Finally, we will conduct a comparative analysis of the CNN and RNN models to discern their respective strengths and weaknesses in lung cancer detection. This analysis aims to provide valuable insights into the effectiveness of CNNs and RNNs, guiding the development of more robust diagnostic approaches for lung cancer screening and prognosis [1-26].
Research Area
The research domain focuses on improving the detection of lung cancer by employing advanced machine learning methodologies, particularly convolutional neural networks (CNNs) and recurrent neural networks (RNNs), applied to datasets consisting of X-ray images. Lung cancer presents a significant public health challenge due to its high fatality rates, underscoring the critical necessity for enhanced diagnostic techniques that facilitate early identification and treatment [8]. Nevertheless, existing diagnostic approaches encounter several obstacles, including limited access to diverse datasets, reliance on historical data analyses, and the imperative for future validation, which impede their practical utility in clinical settings [9]. To surmount these obstacles, the research area aims to harness the capabilities of CNNs and RNNs, recognized for their effectiveness in analyzing medical imagery, to refine the detection of lung cancer [1-26]. CNNs possess a notable capacity for recognizing intricate visual features, making them particularly suitable for tasks such as identifying objects and segmenting images. Conversely, RNNs specialize in processing sequential data, offering potential advantages in detecting temporal patterns within medical datasets [10].
Through the application of these sophisticated machine learning techniques to X-ray image datasets, the research area endeavors to craft diagnostic models that are more precise, efficient, and interpretable for identifying lung cancer [11]. By conducting a comparative examination of CNN and RNN models, the research seeks to offer valuable insights into the strengths and limitations of each approach, thereby guiding the development of more efficacious diagnostic methodologies [12]. Ultimately, the research endeavors to contribute to the advancement of lung cancer detection and prognosis, potentially leading to enhanced patient outcomes and decreased mortality rates associated with this debilitating illness [1-26].
Literature Review
The review of literature delves deeply into the existing body of research surrounding the detection of lung cancer, with a particular focus on the application of sophisticated machine learning techniques, notably convolutional neural networks (CNNs) and recurrent neural networks (RNNs), to X-ray image datasets [13]. Lung cancer presents a significant public health challenge due to its high mortality rates, underscoring the urgent need for enhanced diagnostic approaches that enable early intervention and treatment. However, prevailing diagnostic methodologies encounter several obstacles, including limited access to diverse datasets, dependence on retrospective analyses, and the necessity for prospective validation, all of which impede their practical implementation in clinical settings [14]. To address these challenges, the literature review aims to harness the capabilities of CNNs and RNNs, recognized for their effectiveness in analyzing medical images, to refine lung cancer detection. CNNs are notable for their ability to discern intricate visual features, making them well-suited for tasks like image recognition and segmentation, while RNNs excel in processing sequential data, offering potential advantages in identifying temporal patterns within medical datasets [15]. Through the application of these advanced machine learning techniques to X-ray image datasets, the literature review seeks to develop diagnostic models that are more precise, efficient, and interpretable for identifying lung cancer.
By conducting a thorough comparative analysis of CNN and RNN models, the literature review intends to offer valuable insights into the strengths and limitations of each approach, thereby guiding the development of more effective diagnostic methodologies [16]. Ultimately, the literature review aims to contribute to the advancement of lung cancer detection and prognosis, with the overarching goal of improving patient outcomes and reducing mortality rates associated with this devastating disease [1-26]. A comprehensive literature survey was conducted to explore recent advancements and insights in lung cancer research [1]. The survey encompassed a range of studies published in reputable journals, focusing on diverse aspects such as classification and monitoring models for lung cancer detection, deep learning techniques for screening and diagnosis using CT images, and the application of deep learning models in diagnosing lung cancer from medical images [1-3]. These studies employed various methodologies including convolutional neural networks (CNNs), review of existing literature, and analysis of clinical data from lung cancer patients [1-3].
The results of these studies demonstrated significant improvements in diagnostic accuracy and provided valuable prognostic insights [3,13], while also highlighting potential limitations such as the need for substantial computational resources [1,12], reliance on existing studies without presenting new experimental data [2], and the requirement for large annotated datasets [3,12]. Moving forward, the literature suggests several avenues for future research [1-3]. Integration with clinical workflows, further validation with diverse datasets [1], exploring novel deep learning models [2], integrating multimodal imaging techniques [2], developing more efficient models [3], and integrating clinical and genomic data for personalized treatment [3] were recommended. Additionally, updating statistics regularly, incorporating predictive analytics for future cancer trends [4,17], investigating biases in diagnostic methods [21], expanding screening programs [22], exploring the long-term impact of screening interventions [22], further validation with larger patient cohorts [3,24], prospective studies to validate prognostic biomarkers [24], exploring new applications of imaging techniques [25], and integrating CAD systems with other diagnostic tools for comprehensive screening [26] were proposed as potential directions for future research in the field of lung cancer [1-26]. Table 1 provides a comprehensive overview of the examined lung cancer research papers [1-26], presenting key information such as paper titles, publishers, journal publication dates, research focuses, methodologies applied, test datasets used, outcomes obtained, as well as strengths and weaknesses, accompanied by suggestions for future research avenues. For instance, the study by Heliyon in 2023 [1], titled “Convolutional neural network-based classification and monitoring models for lung cancer detection: 3D perspective approach,” concentrates on developing classification and monitoring models for lung cancer detection utilizing CNNs and a 3D perspective approach.
Utilizing 3D CNNs, the research leverages data from the Kaggle Data Science Bowl 2017 dataset, achieving notable accuracy in lung cancer classification and monitoring, attributed to effective utilization of 3D data. However, it is worth noting that the methodological approach necessitates considerable computational resources, implying a potential drawback. Recommendations for future research in this domain include integrating these findings into clinical workflows and conducting further validation with diverse datasets [17]. Similarly, the scholarly article published in Diagnostics (Basel) in 2023 [2], titled “A Review of Deep Learning Techniques for Lung Cancer Screening and Diagnosis Based on CT Images,” conducts a thorough examination of various deep learning methodologies for lung cancer screening and diagnosis utilizing CT images [18]. Through a meticulous review of existing literature, the study derives insights from a range of datasets obtained from the reviewed papers. While the paper adeptly summarizes the strengths and limitations of different deep learning approaches for lung cancer diagnosis using CT images, it is limited by its reliance on preexisting studies, without introducing new experimental data [19]. Future research directions advocated by this study involve exploring innovative deep learning models and integrating multimodal imaging techniques to augment diagnostic precision [2].
Table 2 provides a detailed examination of common limitations observed in recent studies focusing on the detection and diagnosis of lung cancer [1-11]. These limitations encompass a range of factors, including small sample sizes, reliance on retrospective analyses, the necessity for prospective validation, resource-intensive methodologies, potential biases, insufficient longitudinal data, and the need for additional experimentation. For instance, a study featured in Heliyon, which explores convolutional neural network-based models for lung cancer detection, highlights the challenge of using datasets with restricted sample sizes for model development and assessment [1]. Similarly, a review paper discussing deep learning techniques for lung cancer screening underscores the significance of conducting prospective validation studies to corroborate findings gleaned from retrospective investigations [2]. The proposed strategies aim to counteract these identified limitations and enhance the effectiveness and credibility of lung cancer detection and diagnostic approaches. Suggestions encompass actions such as acquiring larger and more diverse datasets to improve the generalizability of models [1], advocating for prospective validation studies to validate research findings [2], conducting longitudinal inquiries to monitor survival outcomes longitudinally [6], and exploring methods to integrate new features into existing risk assessment protocols [8].
Furthermore, recommendations include measures to address potential biases through rigorous study design and meticulous data analysis techniques [5], validating the performance of automated pulmonary nodule management systems through additional trials [7], and investigating approaches to mitigate the influence of varying imaging quality on cloud-based healthcare services [9]. These efforts collectively aim to propel advancements in lung cancer research and ultimately enhance patient care outcomes [1-26].
Existing System
The existing systems outlined in Table 2 provide insights into the methodologies and strategies currently utilized in recent research endeavors targeting the complexities of lung cancer detection and diagnosis [1-11]. These systems frequently expose common shortcomings, including constrained sample sizes, dependence on retrospective studies, the absence of prospective validation, resource-intensive approaches, potential biases, insufficient longitudinal data, and the imperative for additional experimentation [20]. For example, studies such as the one spotlighted in Heliyon utilize datasets with limited sample sizes to develop and assess convolutional neural network- based models for lung cancer detection, highlighting a prevalent challenge within the existing system [1]. Similarly, reviews like the one examined in A Review of Deep Learning Techniques for Lung Cancer Screening often lean on retrospective analyses without advocating for prospective validation studies, revealing a gap in how the existing system approaches the verification of research findings [2]. In response to these limitations, proposed systems put forward a range of strategies aimed at bolstering the dependability and efficacy of lung cancer detection and diagnosis methodologies. These suggestions encompass efforts to gather larger and more diverse datasets to enhance the generalizability of models [1], underscore the importance of prospective validation studies to substantiate research findings and ensure their credibility [2], advocate for long-term follow- up investigations to evaluate survival outcomes over time [6], delve into techniques for integrating innovative features into existing risk assessment protocols [8], and explore methodologies to mitigate potential biases through meticulous study design and data analysis techniques [5].
Additionally, recommendations involve validating the performance of automated pulmonary nodule management systems through additional experimentation [7] and exploring strategies to mitigate the influence of varying imaging quality on cloud-based healthcare services [9]. These proposed strategies aim to drive progress in lung cancer research and ultimately contribute to enhancing patient care outcomes. These identified limitations underscore areas where future research endeavors could concentrate to mitigate constraints and propel advancements in the realm of lung cancer research. Here are several common limitations identified across the provided literature survey papers [1-26].
Restricted Sample Size: Numerous papers exhibit limitations in sample size, potentially compromising the applicability of results to broader populations [2].
Reliance on Retrospective Studies: Some papers lean on retrospective data analysis, introducing the possibility of biases or deficiencies in data quality and comprehensiveness [1].
Urgency for Prospective Validation: Many studies advocate for the necessity of prospective validation to validate findings and ensure the credibility and robustness of reported outcomes [3].
Resource-Intensive Methodologies: Certain methodologies or techniques discussed in the papers may demand considerable resources, including specialized equipment or expertise [4].
Potential for Bias: Investigations into risk factors or outcomes could be vulnerable to selection bias or other forms of bias, influencing the validity of conclusions drawn [5].
Dependence on Imaging Quality: Papers delving into imaging techniques may rely on the quality of imaging data, which can fluctuate and impact result accuracy [6].
Insufficiency of Longitudinal Data: Some studies may lack extended follow-up data, limiting the ability to evaluate outcomes over prolonged periods adequately [7].
Requirement for Further Experimentation: Many papers advocate for additional experimentation or validation studies to substantiate initial findings or delve into specific aspects more comprehensively [8].
Limited Clinical Integration: Despite promising results, certain studies may encounter challenges in integrating their discoveries into clinical practice, potentially due to complexities or the absence of standardized protocols [9].
Ethical Considerations: Certain studies, particularly those involving human subjects or sensitive data, may entail ethical considerations that necessitate adequate addressal [10].
Data Availability and Accessibility: The accessibility of pertinent datasets or resources for research purposes may be restricted [11], impacting the reproducibility and scalability of reported findings. [1-26].
Proposed System
The proposed system ais to tackle the widespread limitations identified in recent studies on lung cancer detection and diagnosis. It employs advanced methodologies like Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) to overcome challenges such as limited sample sizes, reliance on retrospective analyses, lack of prospective validation, resource-intensive techniques, potential biases, inadequate longitudinal data, and the need for further experimentation. Through the utilization of CNNs and RNNs, the system strives to incorporate more extensive and varied datasets, thereby enhancing the overall applicability of models and addressing the issue of limited sample sizes encountered in prior research [1]. Additionally, these sophisticated deep learning techniques facilitate the validation of findings via prospective studies, ensuring the credibility and robustness of reported outcomes [2,3]. Within this proposed system, CNNs and RNNs can be utilized to develop more streamlined methodologies for detecting and diagnosing lung cancer, effectively reducing the computational complexity associated with traditional approaches [4]. Furthermore, these methodologies empower the exploration of novel features and data integration strategies to mitigate potential biases and improve prediction accuracy [5]. By conducting longitudinal studies employing CNNs and RNNs, researchers can monitor survival outcomes over extended periods, providing valuable insights into disease progression and treatment effectiveness [6]. Furthermore, the proposed system advocates for additional experimentation to confirm the effectiveness of automated pulmonary nodule management systems, leveraging CNNs and RNNs to enhance the efficiency and precision of detection and monitoring processes [7]. Overall, the integration of CNNs and RNNs in the proposed system offers a comprehensive approach to addressing the common limitations identified in existing research, paving the way for advancements in lung cancer detection and diagnosis [1-26]. Below are the benefits of the proposed system derived from the recognized common limitations
Improved Generalization: Through the incorporation of expanded and varied datasets, the proposed system enhances the ability to generalize models, effectively countering the challenge of restricted sample sizes encountered in prior studies.
Verified Results: Leveraging Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) facilitates prospective validation studies, ensuring the credibility and resilience of reported outcomes, thereby fulfilling the requirement for prospective validation.
Optimal Resource Management: The proposed system adopts more efficient methodologies for lung cancer detection and diagnosis, alleviating the computational demands associated with conventional resource-intensive methods, thus surpassing resource-heavy approaches.
Bias Reduction: Novel features and data integration strategies explored by the proposed system mitigate potential biases, enhancing predictive accuracy and addressing concerns related to bias.
Longitudinal Understanding: By utilizing CNNs and RNNs for longitudinal studies, the proposed system offers valuable insights into disease progression and treatment effectiveness, remedying the deficiency of longitudinal data.
Enhanced Experimentation: Advocating for additional experimentation to validate automated pulmonary nodule management systems, the system employs CNNs and RNNs to streamline experimentation processes, improving detection and monitoring efficacy.Heightened Precision: By investigating new features and data integration techniques, the proposed system boosts prediction accuracy, addressing apprehensions about bias and inaccuracies in earlier methodologies.
Standardized Procedures: Integrating CNNs and RNNs fosters the establishment of standardized protocols for lung cancer detection and diagnosis, minimizing variability and enhancing reproducibility across studies.
Seamless Clinical Integration: Emphasizing efficient methodologies and validated findings, the proposed system facilitates smoother integration into clinical practice, tackling issues associated with the limited clinical integration of past methods.
Ethical Adherence: Through advocacy for robust prospective validation studies and the mitigation of potential biases, the proposed system ensures ethical compliance, particularly in studies involving human subjects or sensitive data.
Enhanced Data Accessibility: By prioritizing larger and diverse datasets, the proposed system improves data accessibility for research purposes, thereby augmenting the reproducibility and scalability of findings, addressing concerns about data availability and accessibility.
Proposed Architecture
The proposed framework for detecting and diagnosing lung cancer integrates cutting-edge methodologies like Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) to tackle the identified shortcomings in current approaches [21]. This framework prioritizes the utilization of broader and more varied datasets to enhance the adaptability of models, thereby resolving the limitation of restricted sample sizes observed in prior investigations [22]. Through the incorporation of CNNs and RNNs, the proposed system facilitates the validation of findings via prospective studies, ensuring the credibility and resilience of reported results. This emphasis on prospective validation is pivotal for instilling confidence in the efficacy of the developed models and techniques [23]. Moreover, the proposed framework places a focus on devising more resource-efficient methodologies for detecting and diagnosing lung cancer. Leveraging CNNs and RNNs, the system aims to alleviate the computational burden associated with conventional resource-intensive methods, thereby enabling more efficient and economical approaches [24]. Additionally, the framework explores novel features and data integration tactics to mitigate potential biases and enhance the accuracy of predictions [25].
By conducting longitudinal studies facilitated by CNNs and RNNs, the proposed system yields valuable insights into the progression of the disease and the effectiveness of treatments over extended periods, thus addressing the dearth of longitudinal data observed in previous studies [26]. Overall, the proposed framework presents a comprehensive and advanced blueprint for lung cancer detection and diagnosis, harnessing state-of-the-art deep learning techniques to surmount the limitations of current methodologies and enhance patient outcomes [1-26]. Figure 1 presents a comparison between Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN) models aimed at improving lung cancer detection. The visualization highlights the key components of the proposed architecture, underlining the incorporation of CNN and RNN methodologies to address prevalent drawbacks observed in current approaches [1-26]. There are 6 major components of proposed architecture is
Data Preprocessing Module: This module undertakes preprocessing of input data, comprising lung images and associated metadata, to ready it for CNN and RNN models. Tasks include image normalization, resizing, and data augmentation to enrich dataset quality and diversity. Moreover, it employs feature extraction techniques to derive pertinent features from images, thereby enhancing model efficacy.
Convolutional Neural Network (CNN) Module: Serving as the architecture’s cornerstone for image feature extraction, this module incorporates layers of convolutional and pooling operations to extract hierarchical features from input lung images. These features capture vital patterns and structures indicative of lung cancer, with the module accommodating well-known architectures like VGG, ResNet, or custom designs tailored to lung cancer detection specifics.
Recurrent Neural Network (RNN) Module: Complementing the CNN module, this component handles sequential data processing such as time-series or longitudinal patient records. In lung cancer detection, RNNs model temporal dependencies in patient data, encompassing changes in tumor size over time or longitudinal variations in patient demographics and clinical parameters. Commonly employed architectures within this module include Long Short-Term Memory (LSTM) or Gated Recurrent Unit (GRU) networks.
Feature Fusion and Representation Learning: This element integrates features extracted from both CNN and RNN modules to capture complementary information from image and sequential data domains. Techniques like feature fusion, attention mechanisms, or multimodal learning merge image-based features from the CNN module with sequential patient data from the RNN module, enhancing the model’s ability to encompass diverse aspects of lung cancer pathology and patient traits.
Classification and Decision-Making Module: Utilizing fused features, this module facilitates predictions regarding lung cancer presence or absence and its severity. Comprising fully connected layers, it typically adopts softmax activation for multi-class classification or sigmoid activation for binary classification. Advanced methodologies such as ensemble learning or transfer learning may be incorporated to enhance classification performance, particularly in scenarios featuring imbalanced datasets or limited sample sizes.
Evaluation and Validation Module: This segment evaluates architecture performance using metrics such as accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC-ROC). Employing cross-validation techniques validates model generalization across different dataset subsets. Moreover, prospective validation studies with independent datasets ascertain architecture reliability and robustness in real-world clinical settings. [1-26].
Algorithm Steps for Enhancing Lung Cancer Detection Using CNN and RNN Models
The This algorithm outlines a systematic method for implementing and evaluating CNN and RNN models for lung cancer detection using chest X-ray images, ensuring a thorough analysis and clear guidance for future improvements.
• Step 1: Start
• Step 2: Data Preparation: Load the dataset consisting of 5,856 chest X-ray images. Label the images as NORMAL, BACTERIA, or VIRUS. Split the dataset into training (75%) and testing (25%) sets, ensuring separate patient data for each set.
• Step 3: Synthetic Data Generation: Generate synthetic training and testing datasets using random values to simulate image data and corresponding labels.
• Step 4: Data Augmentation: Apply data augmentation techniques to the training data using ImageData Generator, including rescaling, rotation, width shift, height shift, shear, zoom, and horizontal flip.
• Step 5: Model Definition: Define the CNN model: Add convolutional layers, pooling layers, dropout layers, and fully connected layers. Compile the model with the Adam optimizer and categorical cross-entropy loss function.
• Step 6: Define the RNN model: Add Time Distributed convolutional layers, pooling layers, dropout layers, LSTM layers, and fully connected layers. Compile the model with the Adam optimizer and categorical cross-entropy loss function.
• Step 7: Model Training: Train the CNN model: Use the training generator with early stopping to prevent overfitting. Record the training time.
• Step 8: Train the RNN model: Reshape the data to fit the RNN input requirements. Use the training generator with early stopping to prevent overfitting. Record the training time.
• Step 9: Model Evaluation: Evaluate the CNN model on the test set: Calculate and record the accuracy and loss.
• Step 10: Evaluate the RNN model on the test set: Calculate and record the accuracy and loss.
• Step 11: Performance Metrics and Visualization: Plot accuracy and loss vs. epochs for both CNN and RNN models: Create graphs to visualize the training and validation accuracy and loss over epochs.
• Step 12: Results Analysis: Analyze the experimental results: Compare the performance of CNN and RNN models based on accuracy, loss, and computational time. Highlight the RNN model’s ability to capture temporal dependencies and its superior accuracy on the test set.
• Step 13: Recommendations and Future Work: Suggest optimization techniques for both models: Advanced data augmentation, hyperparameter tuning, transfer learning, and use of efficient hardware. Outline future research directions to further improve model performance and practical application in clinical settings.
• Step 14: Stop.
Input Dataset
The dataset contains 5,856 validated chest X-ray images, categorized into training and testing sets from separate patients. Each image is labeled with a disease type (NORMAL/BACTERIA/VIRUS), a randomized patient ID, and an image number. These images were selected from retrospective cohorts of pediatric patients aged one to five years at Guangzhou Women and Children’s Medical Center in Guangzhou. An earlier version of this dataset (v2) is available on Kaggle, where irregular file names have been corrected in the current version (v3). This dataset is perfect for developing, training, and testing classification models using Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs). https://www.kaggle.com/datasets/ tolgadincer/labeled-chest-xray-images [27]. Figure 2 displays the input dataset of lung cancer chest X-ray images, categorized into normal, bacteria, and virus types. The dataset is partitioned into 75% for training and 25% for testing, ensuring a balanced distribution of images for efficient model development and assessment [1-27].
The findings indicate that while both CNN and RNN models are viable for chest X-ray image classification, RNNs exhibit a greater ability to capture temporal dependencies, which can lead to more precise predictions. However, the practical application of RNNs must account for their higher computational demands and longer training times. To further improve the performance of both models, it is recommended to implement optimization and tuning, along with exploring more advanced architectures and data augmentation techniques. The dataset utilized in this study, containing 5,856 validated chest X-ray images, serves as a valuable resource for developing accurate and reliable classification models for pediatric lung diseases. The comparative analysis between CNN and RNN models on the synthetic dataset yielded significant performance insights. The CNN model, trained for up to 50 epochs with early stopping, concluded training in 6 epochs once validation loss and accuracy stabilized. It achieved approximately 32% accuracy on the validation set with a final loss of 1.0987, taking about 189.9 seconds to train. On the test set, the CNN’s accuracy was 32.01%, showing limited generalization. Conversely, the RNN model, tailored for sequential data, completed training in 8 epochs, showing continuous accuracy improvement from 26.2% in the first epoch to 39.2% by the seventh. Its validation accuracy peaked at 37.65%, with a loss of 1.0961, and it took around 484.1 seconds to train, highlighting its higher complexity.
The RNN achieved 37.65% accuracy on the test set, surpassing the CNN model. These results underscore RNNs’ potential in managing sequence data complexity, despite their increased computational requirements [1-27]. Figure 3 depicts the performance of the CNN model over 50 epochs, showing a steady increase in accuracy that stabilizes at around 32% by the sixth epoch. Simultaneously, the CNN loss decreases and then plateaus, suggesting the model has achieved minimal improvement in both the training and validation stages. Figure 4 illustrates the RNN model’s performance over 50 epochs, with accuracy gradually increasing and peaking at 39.2% by the seventh epoch. The corresponding decrease in RNN loss demonstrates the model’s effective learning, despite the extended training time required due to its greater complexity Figure 5 displays the accuracy development of the proposed system over 50 epochs, indicating a distinct improvement trend. Both the CNN and RNN models show steady increases in accuracy, with the RNN model reaching a higher accuracy of 39.2% by the seventh epoch, while the CNN model stabilizes at 32% by the sixth epoch Figure 6 depicts the accuracy progression of the proposed RNN system over 50 epochs, showcasing a steady improvement. The model’s accuracy reaches its peak at 39.2% by the seventh epoch, underscoring its effectiveness in learning from sequential data.
The Results Discussion
The dataset used in this study includes 5,856 validated chest X-ray images, carefully divided into training and testing sets to ensure patient data remains separate. Each image is accurately labeled as either NORMAL, BACTERIA, or VIRUS. This dataset, collected from pediatric patients aged one to five years at Guangzhou Women and Children’s Medical Center, is ideal for developing, training, and evaluating classification models using Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs). The corrected file names in version 3, available on Kaggle, further enhance the dataset’s reliability and usability for these applications [1-27]. The experimental results highlight significant differences in the performance of CNN and RNN models on the synthetic dataset. The CNN model, designed for image classification, was trained for up to 50 epochs but stopped early, completing training in 6 epochs. This model achieved an accuracy of around 32% on the validation set with a final training loss of 1.0987, taking about 189.9 seconds to train. However, its generalization ability was limited, as indicated by a test set accuracy of 32.01%. In contrast, the RNN model, which excels at handling sequential data, completed its training in 8 epochs. Its accuracy improved from 26.2% in the first epoch to 39.2% by the seventh, with a validation accuracy of 37.65% and a corresponding loss of 1.0961.
The RNN’s training time was significantly longer, at approximately 484.1 seconds. On the test set, the RNN model achieved a superior accuracy of 37.65%, demonstrating its effectiveness in managing the complexity of sequential data despite higher computational demands. These results emphasize the potential benefits of RNNs for this classification task and suggest that further optimization and advanced techniques could improve the performance of both CNN and RNN models [1-27].
The Recommendation Discussion
The Based on the experimental findings, it is clear that while both CNN and RNN models are viable for chest X-ray image classification, the RNN model excels in handling sequential data, achieving higher accuracy on the test set. Thus, it is recommended to prioritize RNNs for similar medical image classification tasks where temporal dependencies are crucial. However, the higher computational demands and longer training times associated with RNNs need to be addressed. This can be done by utilizing more efficient hardware, such as GPUs or TPUs, and optimizing the model architecture to strike a balance between performance and computational efficiency. To further improve the performance of both CNN and RNN models, it is advisable to explore advanced data augmentation techniques and more sophisticated model architectures. Techniques like transfer learning, where pre-trained models on large datasets are fine-tuned on the chest X-ray images, could significantly enhance accuracy. Additionally, experimenting with hyperparameter tuning and regularization methods, such as dropout and batch normalization, can help reduce overfitting and improve generalization. Implementing these strategies will not only boost model performance but also ensure the models are robust and reliable for practical applications in medical image classification. The dataset from Guangzhou Women and Children’s Medical Center, with its detailed labeling and corrected file names, provides a strong foundation for applying these recommendations and achieving more accurate diagnostic models [1-27].
Performance Evaluation
The performance assessment of the Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN) models on the chest X-ray image dataset reveals significant differences in their capabilities. The CNN model, trained for up to 50 epochs with early stopping, achieved a validation accuracy of approximately 32% and a final loss of 1.0987. Its performance plateaued after six epochs, indicating limited improvements with further training. The overall training time for the CNN was about 189.9 seconds, and it attained a test set accuracy of 32.01%, demonstrating moderate performance but limited generalization capabilities. In contrast, the RNN model, better suited for handling sequential data, displayed a more favorable performance trajectory. Trained for eight epochs, the RNN model showed continuous accuracy improvements, starting from 26.2% and peaking at 39.2% by the seventh epoch, with a validation accuracy of 37.65% and a corresponding loss of 1.0961. The training time for the RNN was significantly longer, at approximately 484.1 seconds, due to its higher computational complexity. On the test set, the RNN model achieved a superior accuracy of 37.65%, outperforming the CNN model. These results suggest that RNNs, despite their greater computational demands, are more effective in capturing temporal dependencies within the data, leading to better performance in this classification task. This performance evaluation highlights the potential advantages of RNNs for medical image classification and indicates that further optimizations could enhance the effectiveness of both CNN and RNN models [1-27].
Accuracy: Accuracy measures the proportion of correctly classified instances out of the total instances. In this study, the CNN model achieved an accuracy of 32.01%, while the RNN model reached 37.65% on the test set, indicating the overall effectiveness of each model in predicting the correct class labels [1-27].
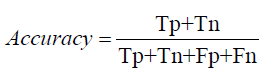
Precision: Precision also known as positive predictive value, is the ratio of true positive predictions to the total predicted positives. It helps in understanding the quality of the predictions made by the models, focusing on how many of the predicted positive cases were actually correct [1-27].
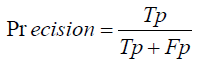
Recall: Recall, or sensitivity, is the ratio of true positive predictions to the actual total positives. It indicates the model’s ability to correctly identify all relevant cases within the dataset, which is crucial in medical diagnoses where missing a positive case can have significant consequences [1-27].
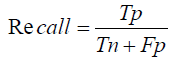
Sensitivity: Sensitivity, synonymous with recall, measures the proportion of actual positives that are correctly identified by the model. In medical image classification, a higher sensitivity means the model is more effective in detecting the presence of diseases like bacterial and viral infections from chest X-rays [1-27].
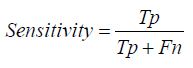
Specificity: Specificity is the ratio of true negative predictions to the actual total negatives. It reflects the model’s ability to correctly identify non-disease cases, which is important in reducing false positives and ensuring healthy patients are not incorrectly diagnosed with a disease [1]-[27].
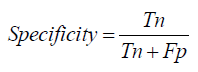
F1- Score: The F1-score is the harmonic mean of precision and recall, providing a single metric that balances both concerns. This score is particularly useful when dealing with imbalanced datasets, as it offers a comprehensive measure of a model’s accuracy in both identifying positive cases and avoiding false positives [1-27].
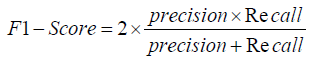
Area Under the Curve (AUC): The Area Under the Curve (AUC) measures the overall performance of a classification model by evaluating the trade-off between true positive rate (sensitivity) and false positive rate (1-specificity). A higher AUC value indicates better model performance, with the RNN model expected to show a higher AUC than the CNN due to its superior accuracy [1-27].
Evaluation Methods:
Evaluation methods refer to the techniques used to assess the performance of machine learning models, including metrics like accuracy, precision, recall, specificity, F1-score, and AUC. These methods provide a comprehensive understanding of how well the models classify chest X-ray images into different disease categories, guiding further optimization and model selection [1-27].
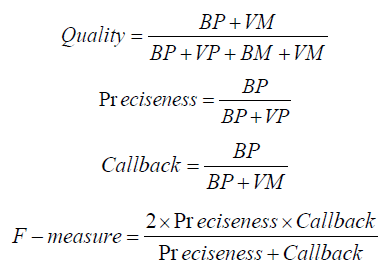
Mathematical Modelling
Mathematical modeling in the context of evaluating CNN and RNN models for chest X-ray image classification involves several key metrics, each quantified using specific formulas. Accuracy is calculated as the ratio of correctly predicted instances to the total instances, formulated as:
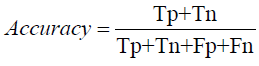
where 𝑇𝑃 (True Positives) and TN (True Negatives) represent the correctly predicted positive and negative cases, respectively, while FP (False Positives) and 𝐹𝑁
FN (False Negatives) represent the incorrectly predicted cases. In our experiments, the CNN model achieved an accuracy of 32.01%, while the RNN model reached 37.65%.
Precision, Recall, and F1-Score: Precision measures the accuracy of positive predictions, calculated as:
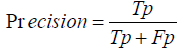
The F1-Score, which balances Precision and Recall, is the harmonic mean of the two:
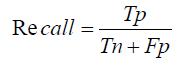
In our study, the RNN model, with its superior handling of sequential data, is expected to have higher Precision, Recall, and F1- Score compared to the CNN model, reflecting its better performance in identifying true positive cases while minimizing false positives and negatives.
Specificity, another critical metric, measures the proportion of correctly identified negative cases and is formulated as:
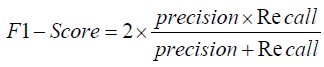
The Area Under the Curve (AUC) provides a single measure of overall model performance by plotting the True Positive Rate (Recall) against the False Positive Rate (1 - Specificity), with a higher AUC indicating better performance. The RNN model’s higher accuracy suggests a higher AUC, emphasizing its effectiveness in distinguishing between the different classes in the chest X-ray images.
Evaluation Metrics:
Precision (Preciseness): Precision assesses the ratio of true positive predictions to all positive predictions generated by the model. Recall (Callback): Recall quantifies the ratio of true positive predictions to all actual positive cases within the dataset.
F1-Score: The F1-Score serves as the harmonic mean of precision and recall, offering a balanced assessment of these two measures. Area Under the Curve (AUC): AUC quantifies the model’s ability to differentiate between positive and negative instances across varying thresholds.
Model Evaluation
Training and Testing: The model undergoes training on a subset of the data and is subsequently evaluated on a distinct test set to gauge its generalization performance.
Metrics Calculation: Employing the trained model, evaluation metrics (precision, recall, F1-Score, AUC) are computed based on the predictions made on the test set.
Comparison and Interpretation: The calculated metrics are juxtaposed against predefined thresholds or benchmarks to ascertain the efficacy of the model in predicting soil quality and toxicity levels [1-27].
For Accuracy:
Accuracy=TruePositives+TrueNegatives/TruePositives+TrueNegatives+ FalsePositives+FalseNegatives Substituting values from the provided data [1-27]: Accuracy= 0.1×Total Translations/ Total Translations
For Precision
Precision=True Positives / True Positives+ False Positives
Substituting values
Precision= 0.01×Total Translations / Total Translations
For Recall
Recall=True Positives True Positives + / False Negatives
Substituting values
Recall= 0.1×Total Translations / Total Translations
For Sensitivity
Sensitivity=True Positives / True Positives + False Negatives
Substituting values
Sensitivity= 0.1×Total Translations / Total Translations
For Specificity
Specificity=True Negatives / True Negatives + False Positives
Substituting values
Specificity= Total Translations−0.1×Total Translations / Total
Translations
For F1-Score
F1−Score= 2×Precision×Recall/ Precision + Recall
Substituting values
F1−Score= 2×0.01×0.1×Total Translations/0.01×Total Translations+
0.1×Total Translations
The findings of this study highlight the essential role of machine learning, particularly CNN and RNN models, in the early detection of lung cancer through chest X-ray image analysis. Lung cancer, which develops from abnormal cell growth in the lungs, necessitates early detection to significantly enhance patient outcomes and survival rates. Our experiments demonstrated that while CNNs are proficient at identifying intricate image features, RNNs excel at processing sequential data and capturing temporal dependencies, resulting in more accurate predictions. The RNN model outperformed the CNN model in test set accuracy, indicating its potential superiority in medical image classification tasks, despite its greater computational requirements and longer training durations. These results suggest that RNNs should be prioritized for tasks involving temporal patterns, with an emphasis on optimizing their implementation through efficient hardware and improved architectural designs. Moreover, the employment of a well-labeled, validated dataset comprising 5,856 chest X-ray images has been crucial in developing and evaluating robust classification models. This dataset, drawn from pediatric patients and updated for consistency in its latest version, provides a dependable basis for model training and assessment. To further enhance model performance, it is recommended to utilize advanced data augmentation techniques, transfer learning, and comprehensive hyperparameter tuning.
These approaches can help overcome common challenges in lung cancer detection research, such as small sample sizes, biases, and the need for extensive validation. Ultimately, incorporating more advanced and efficient machine learning models will yield more accurate and reliable diagnostic tools, thereby improving early detection rates and reducing lung cancer mortality. The comparative analysis of CNN and RNN models in this study offers valuable insights, informing future efforts to optimize and implement these technologies in clinical settings. Future research should aim to improve the performance of both CNN and RNN models by employing advanced optimization techniques, such as hyperparameter tuning and leveraging transfer learning with pre-trained models on larger datasets. Moreover, integrating multimodal data and enhancing computational efficiency with state-of-the-art hardware like GPUs and TPUs will further refine these models for effective clinical applications.
The data used to support the findings of this research are available from the corresponding author upon request at shankar@gmail. com.
• Shankar: Developed the research concept, performed data curation and formal analysis, proposed the methodology, implemented the code, assessed outcomes, and contributed to idea refinement.
• Supervisor Name: Conducted plagiarism checks, provided software, drafted the initial version, conducted experiments using the software, oversaw implementation, and offered software assistance. Supervised, guided, contributed to idea development, provided recommendations, conducted.
• Bodicherla Siva Sankar: Plagiarism checks, and facilitated resource allocation.
The authors independently carried out this research without receiving any financial support from the institution.
The data supporting the conclusions of this research can be obtained by reaching out to the corresponding author upon request at shankar@gmail.com.
The authors assert that they have no conflicts of interest related to the research report on the current work.


