Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Y Zuntz1, David Shemesh1, Sagit Meshulam Derazon2,3, Dean D Ad-El2,3* and David Abookasis1*
Received: June 07, 2024; Published: July 10, 2024
*Corresponding author: David Abookasis, Department of Electrical and Electronics Engineering, Ariel University, Ariel 407000, Israel
Dean D Ad-El, Department of Plastic Surgery and Burns, Rabin Medical Center, Beilinson Campus, Petah Tikva, 49100, Israel, The Sackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv 69978, Israel
DOI: 10.26717/BJSTR.2024.57.009009
In this paper, a hybrid signal processing approach integrating Discrete Wavelet Transform (DWT) and classification by algorithm-based Support Vector Machine (SVM) were used to detect and classify different types of skin cancer from eleven adult patients in a variety of locations on the body. To this aim, a specific optical probe was applied to acquire diffuse light reflectance intensity from the tissue over the visible and near- infrared spectral range (400 - 1000 nm). After outlier detection and removal and Kaiser- Bessel filtering as a preprocessing step on the reflectance intensity spectrum (raw data), DWT analysis with DB4 mother wavelet was used to decompose the raw signal data into 5-levels: D1-D4 and A4 coefficients, representing the optical signal sub-bands. Then, ten features were considered for evaluation, including entropy, kurtosis, skewness, mean, standard deviation, and more. These features were calculated on each DWT coefficient to find the differences in tissue states. After feature extraction and selection, 12 eigenvalues were evaluated as the input of the SVM classifier for classification of normal tissue and different cancer subtypes. Other classifiers including k-nearest neighbor, random forest, and naïve bayesian were also tested and compared. Two different classification algorithms following the above steps have been developed and are describe in this work. Our studies focused on adult patients (ages: 50-90 years old) with diagnoses of malignant melanoma (MM, n=4), basal cell carcinoma (BCC, n=5) and squamous cell carcinoma (SCC, n=2). Measurements were taken first in vivo before surgery, at the site of the lesion and from healthy skin of the same patient, and ex vivo after surgical excision. The results obtained by this work show that both algorithms achieve high performance with more than 95% accuracy. Furthermore, an average area under the curve of 0.99 and a false negative rate of less than 5% are achieved. These results demonstrate the efficacy of the hybrid framework suggested here and support its potential as a suitable automated platform to distinguish between healthy and cancerous skin tissue and to differentiate between cancer types. This could support medical workers, improve the detection rate of cancerous skin lesions, and increase the efficiency of suspicious lesion assessment.
Keywords: Skin Cancer; Reflectance Spectroscopy; Discrete Wavelet Transform; Feature Extraction; Support Vector Machine
Abbreviations: DWT: Discrete Wavelet Transform; SVM: Support Vector Machine; MM: Namely Melanoma; BCC: Basal Cell Carcinoma; SCC: Squamous Cell Carcinoma; DRS: Diffuse Reflectance Spectroscopy; RF: Random Forest; NB: Naïve Bayes; SVM: Support Vector Machine; KNN: k-Nearest Neighbors; MLP: Multilayer Perceptrons; HMM: Hidden Markov Model; AE: Autoencoder
Skin cancer is an anomalous development of skin cells in the epidermis and is the most common form of cancer. It is categorized into two main groups according to their cell origin: namely melanoma (MM) and non-melanoma (BCC: basal cell carcinoma and SCC: squamous cell carcinoma) [1-3]. MM is the most aggressive and dangerous type of common skin cancer due its high capacity to metastasize and is lethal if not promptly diagnosed and treated [4]. BCCs are malignant lesions with low metastasis risk while SCCs can both locally invade, metastasize, and cause death in a subset of patients [5,6]. These types require special attention due to their ability to rapidly grow and possibly result in major disfiguration or even death. Still, they are the most common of all human cancers and a growing health problem worldwide. Over time, many platforms have been suggested for skin cancer detection and diagnosis; each of which possess unique strengths and weaknesses [7-11]. With some of these platforms:
1) The diagnoses are prone to false-positive, or even worse,
false-negative results,
2) Suffer from low penetration depth,
3) Are ineffective at separation between cancer subtypes,
4) Possess a small field of view,
5) Are expensive,
6) Require long screening time, etc.
These drawbacks limit the effectiveness of those techniques as diagnostic tools in the early stage of cancer development. Overcoming these shortcomings, multiple alternative approaches have been suggested that apply a variety of types of hardware together with computational algorithms including artificial intelligence [12-16]. Still, there is an unmet need reliable technique to diagnose skin cancer earlier.
Thanks to the rapid advances in optical technology over decades, its applications in medicine for diagnosis, therapy, and surgery-guidance and specifically for cancer have continually increased and are widely discussed in the literature [17-23]. Among others, optical fiber probe-based diffuse reflectance spectroscopy has been suggested as a tool for early cancer diagnosis via real-time skin cancer monitoring [24-27]. This kind of optical method is considered a popular diagnostic platform due to its ease of operation, high spatial and temporal resolution, relative low cost, noninvasive nature, and lack of risk associated with ionizing radiation. Diffuse reflectance spectroscopy (DRS) depends on the inherent optical properties of tissue (absorption and scattering) and, as such, does not require a contrast agent. Tissue absorption is a function of molecular composition (hemoglobin, melanin, fat, water, and more) while scattering is related to the structure and number of organelles in the cell that have a refractive index different from the surrounding medium [28-30].
In the process of tumor development, as the composition and structure of the tissue changes, light absorption and scattering characteristics also change correspondingly and ultimately affect the reflectance spectrum and its statistical spectral features. In general, the spectrum reflects the chemical, biological, and physical state of the substance and is therefore used in a variety of biomedical applications. Here, in particular, we utilized a DRS configuration based on a single fiber-optic probe composed of seven small fibers: six optical fibers illuminate the tissue, and a single fiber collects diffuse light emitted from the tissue [31-32]. The six illumination fibers are placed around the central collection fiber in a circular manner. We hypothesized that this configuration, in conjunction with the advanced signal processing algorithm and machine learning, could be applied to distinguish between normal and cancer tissue. Furthermore, it could differentiate between cancer types, and serve as a quick, objective, noninvasive screening tool for suspicious skin lesions. Beyond the optical setup is the data processing, where the retrieved bio-optical signals can be analyzed with machine or deep learning approaches to save time and assist health care professionals in the diagnosis task. The use of machine learning algorithms for skin cancer diagnosis is widely studied with a range of detection strategies from simple algorithms to deep learning-based approaches [33-38].
Furthermore, various models are used for classification tasks including decision trees (DT), random forest (RF), naïve Bayes (NB), support vector machine (SVM), k-nearest neighbors (KNN), multilayer perceptrons (MLP), hidden Markov model (HMM), autoencoder (AE), and more. A review of the different methods used in skin cancer classification can be found in Ref. [39]. Here, we chose SVM as the classifier, as they have previously been proven to be very effective. SVM is a supervised learning method that establishes a generalized linear classifier for binary classifications. In general, SVM objective is to find the optimal hyperplane that accurately separates data points belonging to different classes [40-42]. Inspired by the success of the above platforms in different arena, we hypothesize in this study that the use of sophisticated data processing alongside bio- optical methods can improve physician diagnostic accuracy and guide treatment decisions. One of the main challenges in detection of skin cancer subtypes from spectra signals is the detection of the disease with high accuracy in the early stages.
In this paper, with the aim of achieving high accuracy, a hybrid approach wherein a ‘six-around-one’ optical probe combined with discrete wavelet transform decomposition, ten significant features to classify spectra signals, and a machine learning algorithm is implemented for classification of skin cancer samples of 11 adult patients over a wide variety of ages with MM, BCC and SCC on different locations over the body. Before the surgical resection of the cutaneous lesions was conducted, the probe was placed close to the skin area and the reflected light was acquired in the spectral range of 400 to 1000 nm. Then, the probe was placed on a different site on the same patient where the tissue is normal, and measurements were taken again. The patient was then taken to an operating room for local surgery to remove the lesion for histological analysis. During processing, the acquired reflected signal was filtered and outliers were removed as a preprocessing step. Then, discrete wavelet transform (DWT) analysis with a DB4 mother wavelet type was used to decompose the raw signal data into 5-levels (bands). Other kind of mother wavelets were investigated but DB4 was found to give the most accurate results. Ten features were included in the evaluation, including entropy, kurtosis, skewness, mean, standard deviation, and more, were extracted manually on each decomposed signal.
After feature extraction and selection, 12 eigenvalues were evaluated as the input of the SVM classifier for classification of normal tissue and different cancer subtypes. To apply the proposed approach, Matlab software was used for data processing of suspected skin lesions. Figure 1 shows the overall architecture of the proposed technique and steps involved in the classification of optical signal. To increase classification accuracy between cancer types, two different strategies of data analysis were suggested and will be demonstrated (Figures 2 & 3). In general, the accuracy of the classification procedure examines its ability to differentiate cancer from healthy tissue. The results achieved in this study are very promising and show the efficacy of our method to effectively differentiate between cancer subtypes. It should be stress out that although the optical setup and participants are similar to our previous work, 43 the data processing, results and their presentation are completely differing which makes this current research differ in contrast to [43].
Note: Zuntz, et al.
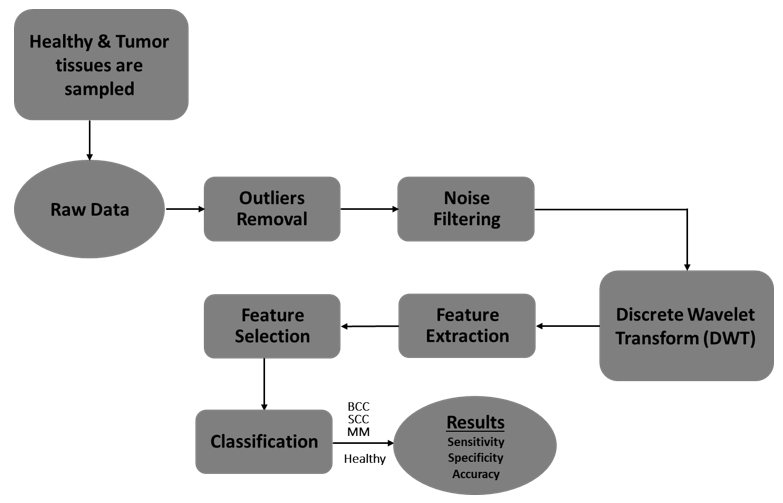
Figure 1: Overall block diagram of the proposed methodology for skin cancer screening and classification.
Note: Zuntz, et al.
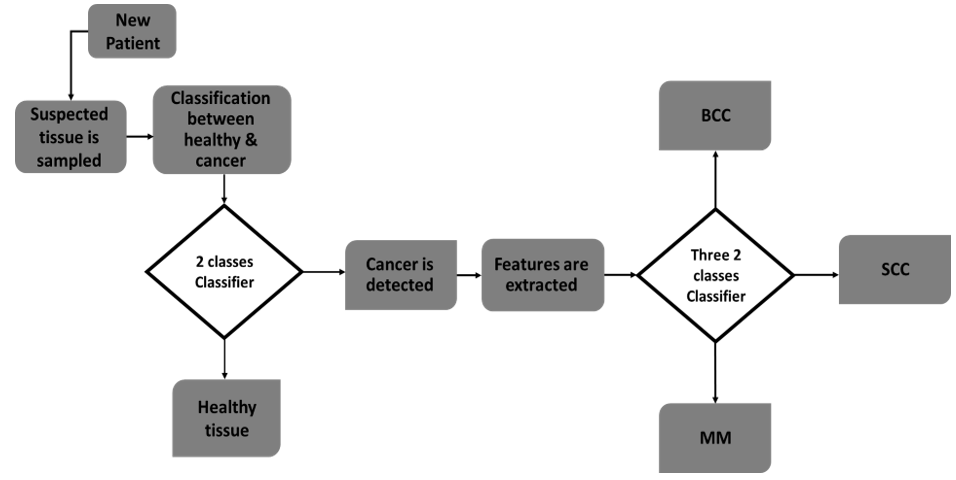
Figure 2: Flowchart of the first approach of classification - SVM was applied in a two- classes classification separating health and cancer tissue and then applied in three-classes classification separating between cancer
Note: Zuntz, et al.
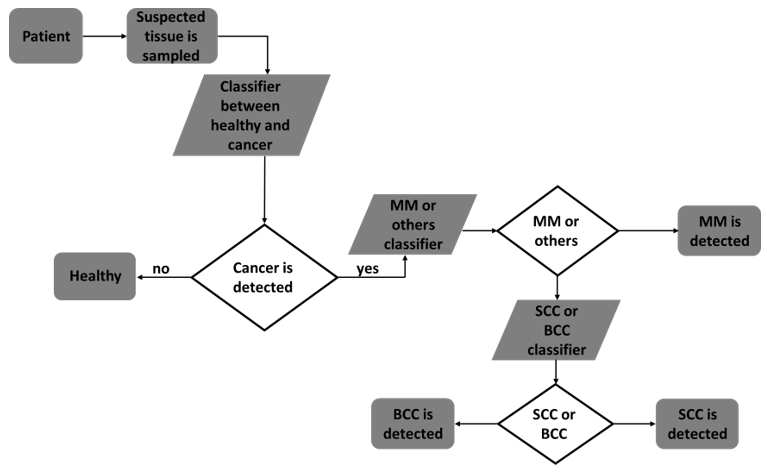
Figure 3: Flowchart of the second approach of classification - after cancer detection SVM was applied in 2-class classification separating MM and the other two cancer types (BCC and SCC). SVM was then applied in a 2-class classification separating BCC and SCC.
The rest of the manuscript is organized as below. Following the introduction in Section 1, the experimental protocol, optical setup, and explains the signal decomposition, feature extraction, and classification used in this work is elaborated in Section 2. In Section 3, the results of the proposed method and the interpretation of the results are displayed. Finally, Section 4 summarizes the paper.
A total of eleven adult patients (male and female, ages 40-90 years old) with observed skin lesions were included in this study. Of the 11 patients, 4 patients were diagnosed with MM, 5 with BCC, and 2 with SCC. Samples included skin excisions from the face, scalp, neck, leg, forehead, chest, and back. Informed consent was obtained from all participants and clinical information was collected. Suspected lesions were identified by a clinician and their diagnoses were confirmed through histological analysis following a biopsy. Prior to surgery, the spectral reflectance of tumor tissue and adjacent normal healthy tissue was acquired and processed off-line. Following surgery, the lesion was rinsed and sent to the pathology laboratory unit in the hospital for analysis according to the standard pathology workflow for tissue processing and histopathological analyses. Pathological results were obtained following 3 - 7 days. This work was carried out in accordance with all ethical guidelines, approved by the internal review board (Helsinki committee) at Rabin Medical Center (study number 0494-21-RMC).
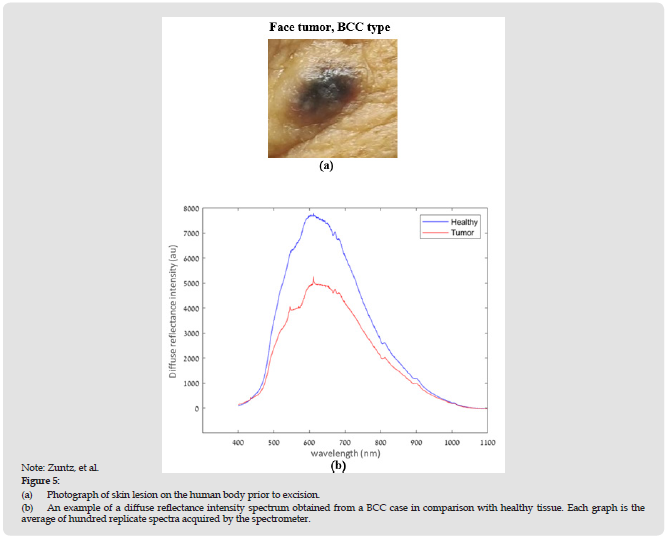
Figure 4 presents a schematic illustration of the setup used in this study. As shown, the system consisted of a wideband light source (Thorlabs, SLS201L), a single fiber-optic probe (Ocean Optics, R200- 7), a USB fiber-optic spectrometer (AvaSpec- ULS4096CL-EVO, Avantes), and a computer (Lenovo G570). During the experiments, the probe was held gently and closely perpendicular to the middle of the tumor. The probe was not in contact with the tissue being tested (5 mm distance) to avoid the need for cleaning and disinfection of the probe. Representative close-up photographic images of the lesion on the human body prior to excision are presented in Figures 5a. The spectrometer operates in the spectral range from 200 to 1100 nm with a spectral resolution of 0.05 nm and is connected to the computer. The probe, which guides the light from the light source toward the tissue, contains six illumination fibers arranged around the central single collection fiber, which transmits the reflected light emanating from the tissue back to the spectrometer [44]. Each set of data acquisition involved the measurement of one hundred replicate spectra (acquired over 13 s) at one position which were co-added to a single average spectrum and used for further analysis. Measurements were taken at the site of the lesion and from healthy skin of the same patient. An example of diffuse reflectance intensity spectrum obtained from a representative BCC case as compared to healthy tissue is shown in Figure 5b. The entire setup was controlled by Matlab software (The Mathorks Inc., MA, USA) which was also used for data processing.
Note: Zuntz, et al.
Figure 4: Schematic diagram of the optical system utilizing a bifurcated fiber containing six irradiation fibers around one collection fiber.

Note: Zuntz, et al.
Figure 5:
(a) Photograph of skin lesion on the human body prior to excision.
(b) An example of a diffuse reflectance intensity spectrum obtained from a BCC case in comparison with healthy tissue. Each graph is the average of hundred replicate spectra acquired by the spectrometer.
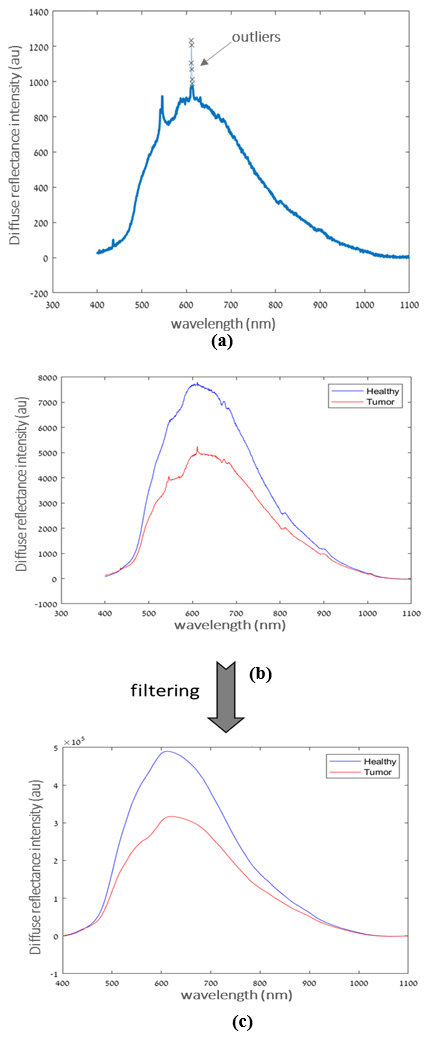
A schematic diagram of the framework is depicted in Figure 1. The first stage consists of a preprocessing chain including detecting and removing outliers and signal filtering to compensate for the effect of environmental conditions and for the system response. In order to prevent errors and uncertainties in the acquired measurement, spectral outliers were identified by the following criterion of: Iraw(λi) ≥ Iavg ± 1.96 ×σ where Iraw(λi) represents the reflectance intensity at the specific wavelength, λi , Iavg is the average of neighboring intensity around λi, and σ the standard deviation of the mean; if identified as an outlier, the intensity at the specific wavelength was removed from the spectra. An example of the detection of six outliers at the wavelength 605 nm is demonstrated in Figure 6a. A variety of FIR filters such as Rectangular, Hann, Hamming, and Kaiser window techniques were tested for noise reduction (denoising) following outlier elimination [45]. After comparing and analyzing the performance of each filter, the Kaiser window order 64 with β=0.5 was found to give the best results. A sample spectrum of cancer (BCC case) and healthy tissue before and after Kaiser filtering method is presented in Figures 6b & 6c, respectively. After the data was denoised, the wavelet transforms method was used to decompose the signal into different components and wavelet coefficients were obtained with a base wavelet function depending on the scale and shift value.
Note: Zuntz, et al.
Figure 6:
(a) Example of the detection of six outliers at 605 nm.
(b) Spectrum profile of cancer and normal tissue before and
(c) After Kaiser filtering.
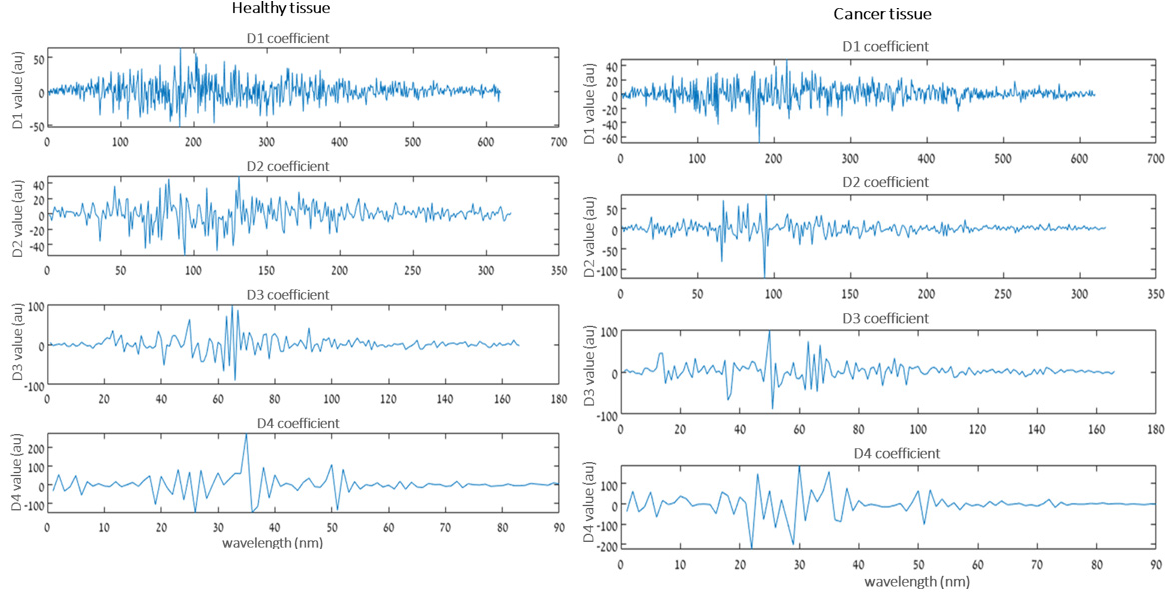
Discreate wavelet transform (DWT) is one type of wavelet analysis that is used here for decomposition. Generally, DWT is performed by repeated filtering of the input signal with a pair of low and high pass filters. The coefficients corresponding to the low pass filter are called approximation and similarly, high pass filtered coefficients are called detailed. Furthermore, the approximation coefficient is consequently divided into new approximation and detailed coefficients. This decomposition process is carried out until the required frequency response is achieved from the given input signal [46,47]. After many experiments, it was finally determined that our signal is decomposed into five levels of wavelet by using Daubechies wavelet function (DB4); the resulting five coefficients of DWT were used to represent the optical signal sub-band as shown by the example in Figure 2. After decomposition, a set of ten common statistical metrics of different orders and non-statistical features were calculated to assess the optical signal corresponding to each sub-band to find the difference in cancer states. The features include the standard deviation, mean, variance, median, max, min, shannon entropy, skewness, kurtosis, and power spectrum density. Ten features were extracted for each of the five subband signals and therefore a total of 50 features were obtained and served as input to the classifier. The classifier directs the classification of a new observation to a particular category.
In search of the best classifier for the proposed data, five classifiers namely random forest, naïve bayes, support vector machine, k-nearest neighbors, and decision trees were tested. In this work, three types of SVM with Gaussian / Quadratic / Cubic as the kernel function were utilized to improve classification results further. SVM is the most common classifier used in science and engineering, as it utilizes non-linear transformation to make an appropriate decision. It is a supervised learning method that aims to find an optimal separating hyperplane with the maximum margin to separate two classes in the feature space [48]. The vectors that define the hyperplane are called the support vectors. In the first approach of classification, SVM was applied in 2-class classification separating the healthy control from the cancer tissue and then applied in 3-class classification separating between cancer subtypes as described in Figure 6. In the second approach, after cancer detection SVM was applied in 2-class classification separating MM and the other two cancer types (BCC and SCC). Then, SVM was applied again in 2-class classification separating BCC and SCC as shown by the block diagram in Figure 7. The performance of these two approaches will be presented in the results section.
Note: Zuntz, et al.
Figure 7: Example of five decomposition coefficients of DWT representing the optical signal sub-band for healthy and cancer (BCC case, Figure 2b) tissue.
We evaluated the performance of the classification using four commonly used parameters for a binary classification task including the area under the curve (AUC), accuracy, sensitivity, and specificity. AUC measures the classifier’s ability and is employed as a ROC curve summary. These quantities are defined in the following equations by four classification values [49,50]

TN = True Negative, TP = True Positive, FP = False Positive, FN = False Negative. TP represents the number of diseased cancers actually predicted by the classifier as diseased and FP represents the number of diseased predicted as healthy. TN is defined as a healthy sample predicted healthy, and FP is the number of mistakenly predicted healthy as diseased. The matrix based on the classification results is shown in Table 1. The data was divided into calibration (training) and validation (testing) for the performance evaluation of the classifiers using the k-fold Cross Validation (CV) approach. In the k-fold CV, the data set is split into k folds which k-1 folds is used for training and one-fold for validation. This procedure is performed for all parts of the data, yielding the results of each fold. The classifier’s overall performance is then assessed by averaging all of the folds. In this study, the performances of the classifiers were evaluated using 5-fold CV. Selection of appropriate feature plays an important role in signal processing. Before classifying the signal, the extracted feature must be selected accordingly to provide the best result, because selection of the wrong features may give poor results even when applying the best classifier.
The study was carried out in two different ways in terms of feature selection and classification approach as shown in Figures 2 & 3. Common to these approaches is the way the dataset was split. As mentioned in subsections 2.1 and 2.2, we have eleven subjects and one hundred samples were acquired from each subject, resulting in total of one thousand hundred samples. This is not the ideal situation given the small number of patients (n =11), but it does give some indication of the viability of our approach. During experiments, this data set (n =1100) is split as 80% for training data and 20% as testing data (validation), randomly. The confusion matrix, which measures the degree of deviation between the labeled experimental outcomes and true ones, of the classification results of the first approach using the Kurtosis of D2 and D4 DWT coefficients as feature vectors and classification scatter map with a quadratic SVM classifier are shown in Figures 8a & 8b, respectively. Specifically, the scatter plot of the classifier includes a marking for the top 50 support vectors to help visualize the decision surface that classifies the observations in the hyperplanes. The confusion matrix is a result of using k-fold cross-validation mentioned in sub-section 2.4. Performance results of accuracy, TP, and FN parameters in classifying cancer subtypes are given in Figure 8c.
Note: Zuntz, et al.
Figure 8:
(a) Confusion matrix of the first classification approach using the Kurtosis of D2 and D4 DWT coefficients. The matrix is a result of using k-fold cross-validation mentioned in sub-section 2.4.
(b) Classification scatter map with a quadratic SVM classifier. The scatter plot includes a marking for the top 50 support vectors to help visualize the decision surface that classifies the observations in the hyperplanes.
(c) Performance results of accuracy, TP, and FN parameters in classifying cancer subtypes

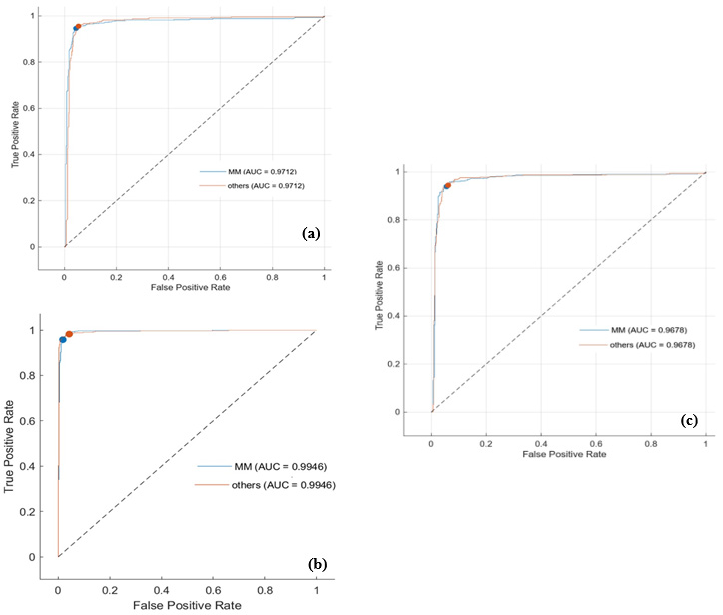
The recognition accuracies of BCC, MM, and SCC are 96.7%, 96.5%, and 97.9%, respectively with overall low FN rate of less than 3.5%. The confusion matrices indicate that our approach achieved higher scores on true positives indicating high sensitivity, which is crucial in the field of medicine, especially when the diagnosis is automated. For further illustration and clarification of results, receiver operating characteristic (ROC) curves based on SVM quadratic kernel are depicted in Figure 9. The larger the TPR, the higher the diagnostic accuracy; The smaller the FPR, the lower the misjudgment rate. The value of ROC ranges from 0.5 to 1, in which 0.5 indicates the worst classifier and 1 indicates the most robust classifier. Area under curve (AUC) is given; the higher the AUC value the better the model’s performance at distinguishing between classes. As depicted, the proposed approach is closer to the point in the upper left corner of the ROC space, indicating that its diagnostic performance is better. It can be seen that the proposed method performed well in the classification of all types, and specifically of SCC with AUC = 0.9976.
Note: Zuntz, et al.
In a second approach considered in this study (Figure 7), the resultant confusion matrix using the average of Shannon of D4 and the minimum of Shannon D4 coefficients as feature vectors and classification scatter map are shown in Figures 10a & 10b, respectively. Performance results of accuracy, TP, and FN parameters in classifying cancer subtypes are given in Figure 10c. The recognition accuracies of MM, and others (BCC and SCC) reached about 94.8% and 95.1%, respectively, with a low FN rate of less than 5.2%. Following final classification for the others, 100% accuracy rates were achieved with maximum TPR as presented in Figure 11. As the figure shows, indeed performance accuracy for MM is about the same here as with the first approach, with the current approach accuracy for BCC and SCC increase up to 100% showing that all the BCC / SCC types were detected correctly. The attained ROC curve based on SVM quadratic kernel is depicted in Figure 12. After obtaining the above results, a comparison of the performance of both classification approaches were conducted using six different classifiers (Fine Tree, Wide Neural Network, Fine KNN, Ensemble-Bagged Trees, SVM - Quadratic Kernel, and Naive Bayes); results are shown in Tables 2 & 3 for both approaches, respectively. Based on the results shown in the tables, the highest accuracy is achieved with Ensemble-Bagged Trees (EBT) classifier with an average accuracy of 98%.
Note: Zuntz, et al.
Note: Zuntz, et al.
Figure 11: Continuation with Figure 8, second classification approach between SCC to BCC. Here, the average of Shannon average of D1 and Kurtosis of D3 coefficients, respectively as feature vectors is used.

Note: Zuntz, et al.
Figure 12: Comparison of ROC performance obtained from classification by the second approach between MM to others (BCC and SCC) using
(a) SVM and Ensemble - Bagged tree.
(b) ROC curve between BCC and
(c) SCC using Quadratic SVM classifier.
In both approaches, the Fine KNN has the lowest performance measure of any comparison method. For further illustration and completeness of presentations, ROC graphs for the different classification classifiers are shown in Figures 13a & 13b for the first and second approach, respectively. Finally, ROC performance for the BCC and SCC in the second approach is shown in Figure 13c. With the results presented above it can be concluded that our approach can be used to detect and classify different types of skin cancer with high accuracy and overall may be adapted to the detection of a variety of other c.
Note: Zuntz, et al.
Figure 13:
(a) ROC curves obtained from different classifiers for the first and
(b) Second approaches
(c) ROC curve of BCC and SCC following the second approach.

This study presented a hybrid pipeline methodology combining discrete wavelet transform and machine learning from broad wavelength range (VIS-NIR) diffuse light reflectance spectra for the discrimination of cancerous from healthy tissue and classification of cancer subtypes. Two different strategies of data analysis were suggested and compared based on ROC performance through a series of six different classifier techniques. During this research, multiple extracted features were used to increase the screening between skin cancer subtypes: SCC, BCC and MM. After decomposition, a set of ten statistical moments and non-statistical features were used to help distinguish cancer states. We demonstrate our approach on cancerous lesions obtained from eleven patients over different parts of the body as compared to healthy skin tissue on each patient. Six different ML methods were carried out, and these models were trained and tested using a 1100 data set. The performance of the proposed algorithm has been validated by measuring various parameters including sensitivity, accuracy, and more. According to our results, we demonstrate that the EBT (ensemble bagged tree) method achieved the highest classification accuracy of at least 98%, with an AUC of 0.99, compared to other classifiers thus underscoring the validity and feasibility of the proposed method for the identification of skin cancer.
Overall, this study yielded excellent results in differentiating normal and cancer tissues in human experiments which has the potential to improve both patient survival and outcome. We believe that our approach can be used as an efficient scheme to assist clinicians for automatic, real-time human skin cancer detection and classification and will inspire further development improving classification and accuracy. Along the same line, we plan to implement this approach during hyperspectral imaging procedures of tissue examination which might be beneficial for the skin cancer diagnosis and disease types.17, [51- 53] One main limitation of this study is the small number of patients and therefore a study with a larger patient sample should be carried out in the future to allow verification and reproducibility of the reported results. To the best of the author’s knowledge, we are the first group to suggest such a skin cancer detection method.


