Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Fakoya A, Ogunlade O A and Olusola A O*
Received: May 16, 2024; Published: June 04, 2024
*Corresponding author: Olusola A O, Department of Biochemistry, Faculty of Sciences, Adekunle Ajasin University, Akungba Akoko, Ondo State, Nigeria
DOI: 10.26717/BJSTR.2024.56.008915
Corona Viruses (CoVs), family of viruses that cause intestinal and respiratory- mild colds illnesses in humans and animals. The emergence of the severe acute respiratory syndrome (SARS) epidemic in China in 2002- 2003 and the Middle East respiratory syndrome (MERS) was a treat to the world. HCoVs have been linked with major outbreaks of human fatal pneumonia since the beginning of the 21st century. Buchholzia coriacea (BC) plant is useful in folk medicine for the treatment of a wide range of diseases due to its secondary metabolites. In this research work, the in-silico study of BC terpenoid compounds was evaluated for their inhibitory potential against SARS spike protein receptor binding pocket. Terpenoid compounds were extracted from BC seed and the Gas Chromatography-Mass Spectrophotometer (GC-MS) analysis showed that it contained; rutin, phytic acid, caffeic acid, flavone, ferulic acid, gallic acid and phenol. These compounds were then docked with SARS spike protein receptor binding domain. Molecular docking revealed that they had high binding affinities compared to the standard drug remdesivir having the binding affinity (-4.740 kcal/mol). The ADME properties of the terpenoids from BC were evaluated to explain their pharmacokinetic and the results revealed that caffeic acid, gallic acid and ferulic acid could therefore serve as lead compounds in the treatment of SARS-COV.
Keywords: Terpenoid; Inhibitory; Corona; Affinities; Spike Protein
Abbreviations: CoVs: Corona Viruses; SARS: Severe Acute Respiratory Syndrome; MERS: Middle East Respiratory Syndrome
Human Corona Viruses
Corona viruses (CoVs), a family of viruses that cause respiratory and intestinal illnesses in humans and animals (Cui et al. [1]). They usually cause mild colds in people but the emergence of the severe acute respiratory syndrome (SARS) epidemic in China in 2002-2003 and the Middle East respiratory syndrome (MERS) on the Arabian Peninsula in 2012 show they can also cause severe disease (Cui et al. [1]). Today, HCoVs are well known for rapid evolution due to high nucleotide substitution and recombination rate (Vijgen et al. [2]). HCoVs have been appeared periodically in deferent places around the world and linked with major outbreaks of human fatal pneumonia since the beginning of the 21st century (Wu et al. [3]). First CoV outbreak as severe acute respiratory syndrome corona virus (SARS-CoV) started in November, 2002 at Foshan, China (Ge et al. [4]) and later turned into global infection in 2003 with a lethal rate of 10% worldwide (Lee et al. [5]). Following one decade, second HCoV pandemic was caused by Middle East respiratory syndrome corona virus (MERS-CoV), originated in June, 2012 at Jeddah, Saudi Arabia (Ge et al. [4]), with a global fatality rate of 35% (de Groot et al. [6]). Recent third major HCoV explosion, occurred in December, 2019 at Wuhan province of China, caused by highly homologous novel strain of SARS-CoV, classified as severe acute respiratory syndrome corona virus-2 (SARS-CoV-2); designated the infection as COVID-19 (Corona Virus Disease 2019) pandemic (Zhu et al. [7]). These HCoVs outbreaks are classified as continuous threat to humans and world economy because of their unpredicted emergence, rapid and easy proliferation that lead to catastrophic consequences (Zhu et al. [7]).
The virus and its host shared a complex relationship, including numerous viral and host factors, for initiation of viral infection; subsequently in potential pathogenesis (Lim et al. [8]). As intracellular obligate parasites, viruses have also advanced various strategies to hi- jack host-cell machineries (Lim et al. [8]). For HCoVs, there is no potential therapeutic agent such as drug or vaccine; thus, strict implementation of high vigilance such as for SARS-CoV-2 has been recommended for prevention and to check the infection (Cheng et al. [9]). Researchers and Authorities (WHO, CDC Atlanta and others) across the globe are working to combat the current on-going SARS-CoV-2 outbreaks, and identifying the possible origin of this novel virus to develop effective therapeutics (Cohen, et al. [10-12]). Therefore, identifying the route of origin and pathogenesis of major pathogenic HCoVs may provide cognizance in the development of potential therapeutics.
These viruses have been confronted on several occasions with prerequisite to classify a newly emerged virus associated to a severe or even fatal disease in humans under existing genera or a new species (Gorbalenya et al. [13]). In this context, current classification distributed 39 species of CoVs in 27 subgenera, 5 genera, and 2 subfamilies that categorized under family Coronaviridae, suborder Cornidovirineae, order Nidovirales, and realm Riboviria (Gorbalenya et al. [13,14]). Herein, HCoVs are categorized under subfamily Coronavirinae of family Coronaviridae, are genotypically and serologically alienated into four major genera; AlphaCoV, BetaCoV, GammaCoV, and DeltaCoV, by International Committee for Taxonomy of Viruses (Wu et al. [3]). Remdesivir, a prodrug of an adenosine nucleotide analogue, is an antiviral agent with broad-spectrum activity against viruses from several families (Brown, et al. [15]). Remdesivir was previously under development for the treatment of Ebola virus disease in the wake of the 2014-2016 Ebola outbreaks in West Africa. While it was a promising therapeutic agent for Ebola virus disease in preclinical studies (Warren, et al. [16]), monoclonal antibodies out performed remdesivir in a phase III clinical trial (Mulangu, et al. [17]) and remdesivir is no longer being developed in this indication. The antiviral activity of remdesivir against corona viruses, however, has rendered the drug of great interest during the current global pandemic.
The novel corona virus disease 2019 (COVID-19), caused by severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) infection, was first reported in Wuhan, China, in December, 2019. The World Health Organization declared COVID-19 a Public Health Emergency of International Concern on 30 January, 2020 and a pandemic on 11 March, 2020 spurring an international effort to rapidly identify treatments that might ease the burden on health care systems. Phase III trials of remdesivir in COVID-19 were initiated as early as February, 2020 (WHO, 2020). Based on data from the multinational phase III ACTT-1 and SIMPLE-severe trials, remdesivir received an emergency use authorization in the USA on 1 May, 2020, and a special approval for emergency use in Japan on 7 May, 2020. Remdesivir received its first conditional approval for use in patients with severe COVID-19 in Taiwan in late May, 2020 with this conditional approval requiring the pharmaceutical company to implement a risk management plan to ensure safety.
Buchholzia. Coriacea Plant
Buchholzia. coriacea was named after R. W. Buchholz who collected plants in Cameroon in the late 19th century (Anie, et al. [18]). It belongs to the Capparaceae family. The seed of B. coriacea has medicinal values. These seeds gave the plants its common name of “wonderful kola” because of its usage in traditional medicine. The seeds are covered in a purple aril which is chewed in Ivory Coast and has a sharp pungent taste. (Burkill [19]) carried out a research on the medical uses of this plant in different parts of Africa. It has been used for years to meet a variety of illnesses; since it has been used continually over many generations it is likely that wonderful kola actually has an effect against illnesses. As a result of its supposed broad-spectrum activity, there is need to conduct studies on potential utilization of wonderful kola in foods. This research is aimed at studying the molecular interaction between secondary metabolites analyzed from the terpenoids of B. coriacea, and SARS spike protein receptor binding domain using Schrödinger Suite and also to evaluate the ADME analysis of the terpenoids of B. coriacea using Qikprop.
Extraction of Terpenoid from Buchholzia Coriacea
The method described by (Jiang et al., 2016) was employed for the extraction of terpenoid from the seed of Buchholzia coriacea. The powdered seed were soaked in n-hexane for 72 hours at room temperature and filtered. The filtrate was concentrated in a rotary evaporator at 50ºC. Terpenoids were extracted from the crude by 85:15 v/v n-hexane, ethyl acetate, and silica gel (pore size 230–400 mesh, Sigma-Aldrich catalog number: 227196). Agilent 6890a GC equipped with an HP5-MS column (30 m × 0.250 mm × 0.25 μm) coupled to an Agilent 5975C Mass Spectral Detector or Flame Ionization Detector (FID) was employed for the characterization of the terpenoids.
Gas Chromatography-Mass Spectrophotometer (GC/MS)
The sample was injected into Gas Chromatography (GC) in a port which was heated up to 3000C where the material then volatilized. The column was wound within a special oven which controls temperatures from -200 to 3200. The column surface is coated with a material which will separate the various chemical compounds in the sample based on size and/or polarity. Sample components that are more volatile and smaller in size will travel through the column faster than others; hence, gaseous components are separated as they flow through the column into the mass spectrophotometer. The mass spectrum is used to identify the components by comparing each to reference libraries of over 275,000 unique spectra. To quantify compounds within the analyzed sample, analysts establish a standard curve of known concentrations of each material.
Ligand Library Generation and Preparation
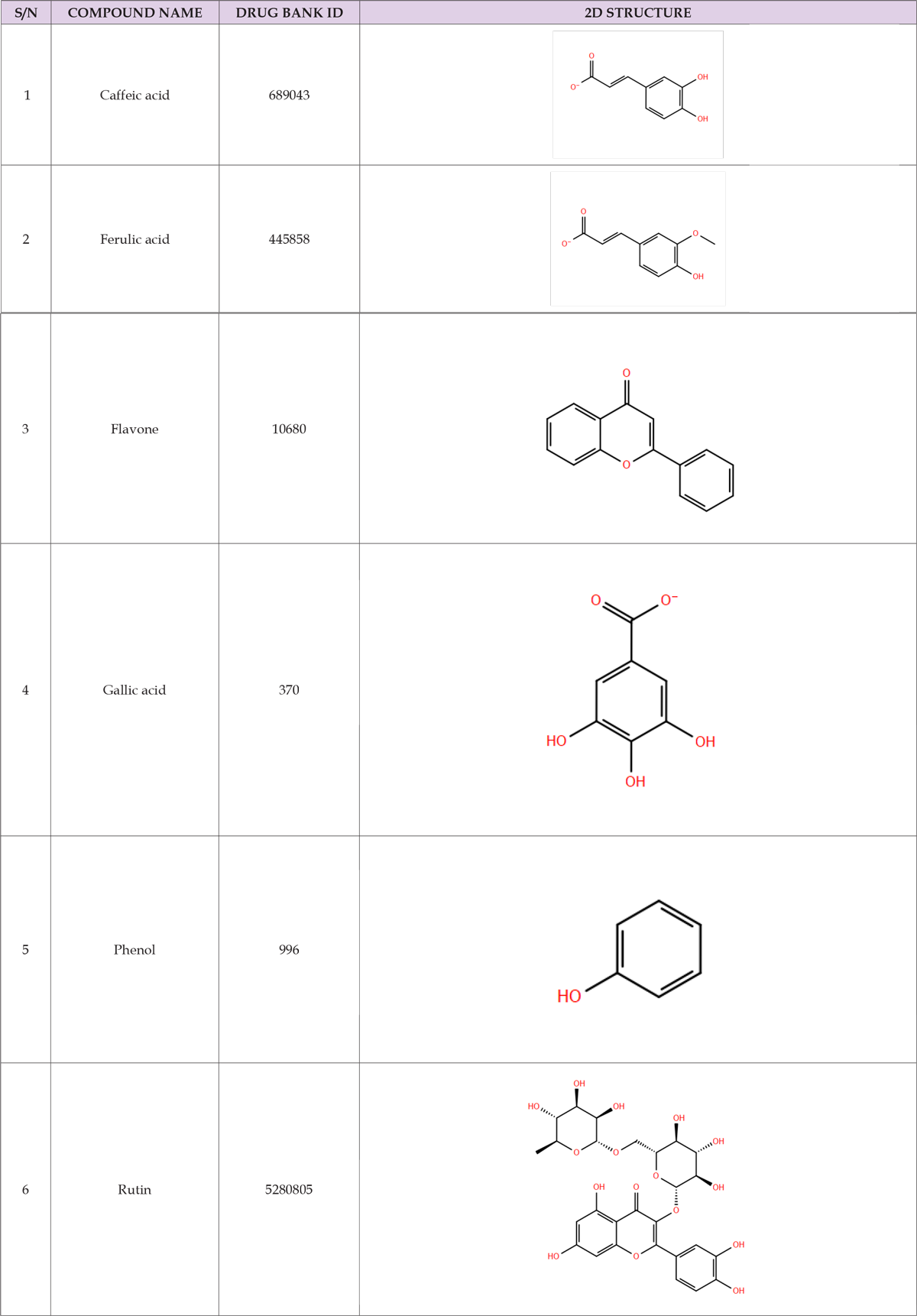
Structures of Terpenoid compounds (two dimensional) Caffeic acid, Ferulic acid, Flavone, Gallic acid, Phenol, and Rutin were mined from PubChem online database (Kim et al., 2016) in sdf format and was prepared using ligprep tool (Schrödinger, 2021), (Using Epik at pH 7.0 with OPLS3 force field). Maestro, Schrodinger Suite 2021 (Schrödinger, 2021). A maximum of 32 possible stereoisomers per ligand were obtained.
Target Retrieval and Protein Preparation
X-ray crystallographic structure of SARS spike protein receptor binding domain (2GHV) complexed with an inhibitor as co-crystallized ligand at the active site (RNA 960) (PDB ID: 2GHV) (Asthana et al., 2014) was retrieved from Protein Data Bank (Berman et al., 2000). It was selected as a result of the presence of an inhibitor as ligand at the active site of the protein (David et al., 2018). The retrieved protein was viewed and prepared using Maestro 12.5 (Schrödinger, 2021), Protein preparation wizard, optimized at pH 7.0 and minimized using OPLS3 as force field. Charges and bond orders were assigned and water molecules were deleted. Hydrogen was added to the heavy atoms.
Receptor Grid Generation
Receptor grid shows the area between the ligand and the protein where interaction takes place. For Glide docking, grids were generated by using OPLS-3 force field by keeping the van-der Waals scaling factor of 1.0 and charge cut-off value of 0.25. A box was generated to each direction with 14 Å × 14 Å × 14 Å for docking experiments.
Extra Precision (XP) Ligand Docking
XP ligand docking was performed rather than SP docking because XP is better than SP in scoring function and it also predicts the false positive results (Friesner et al., 2006). This docking was performed in Glide of Schrödinger Maestro v9.4 (Friesner et al., 2004). Final result of docking can be found as glide score by energy minimization. For docking, van-der Waals scaling factor was set to 0.85 and 0.15 for ligand compounds and partial charges cut-off value was fixed at -10.0 kcal/mole. The lowest glide score containing compounds were then subjected to MM-GBSA analysis for binding free energy calculation and best poses were recorded for every ligand compounds.
Prime MM-GBSA
Binding free energy calculation was also carried out for the protein ligand complexes. MM-GBSA is a combined method for binding free energy calculation which was used in this experiment that accumulates OPLSAA molecular mechanics energies (EMM), an SGB solvation model for polar solvation (GSGB), and a non-polar solvation term (GNP) composed of the non-polar solvent accessible surface area and van-der Waals interactions (Rastelli et al., 2010). The best poses from the Glide score were used for binding free energy calculation. The total free energy of binding: ΔGbind = Gcomplex – (Gprotein + Gligand), where G = EMM + GSGB + GNP
Ligand based ADME Analysis
Analysis of physiological descriptors of a compound such as adsorption, distribution, metabolism and excretion behaviour of the ligand compounds ADME analysis was done in QikProp module of Schrodinger (Natarajan et al., 2015). It also predicts the physico- chemical nature of the compounds as well as their pharmacokinetics properties. In this study, we used the Qikprop 3.2 module of Schrodinger (Sharma et al., 2011). There are also several other descriptors also analyzed such as Predicted IC50 for blocking HERG K+channel in-vitro, predicted octanol or water partition coefficient [log P(o/w)], number of hydrogen bond acceptors (HBA), number of hydrogen bond donors (HBD), predicted aqueous solubility (log s), solvent-accessible surface area (SASA), skin permeability (logKp), MDCK cell permeability (MDCK), binding to human serum albumin (logKhsa), blood-brain partition coefficient (logBB), percentage human oral absorption rate (Table 1).
Table 1: 2D structure of terpenoids from buchholzia coriacea extract obtained from NCBI PUBCHEM and EMBL-EBI database.
(Tables 2-4) and (Figures 1-7).
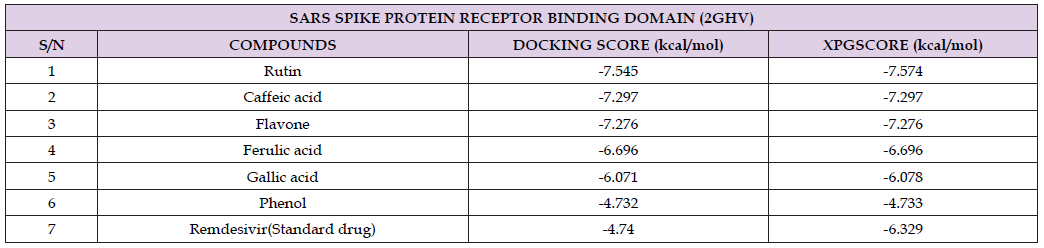
Table 2: Docking score of SARS spike protein receptor binding domain (2GHV) with the secondary metabolites analysed from B. coriacea and remdesivir as the standard drug.
Note: WHERE;
Total solvent accessible surface area, SASA = 300.0-1000.0
Hydrogen bonds donor, HB donor = 0.0-6.0
Hydrogen bonds acceptor, HB acceptor = 2.0-20.0
Predicted IC50 value for blockage of HERG K+ channels, QPlogHERG = Concern below -5
Predicted qualitative human oral absorption, HOA (%) = >80% is high, <25% is poor
Predicted blood/brain partition coefficient, QPlogBB = -3.0-1.2
Predicted value for serum protein binding QPlogKHsa
Predicted octanol/water partition coefficient, QPlog Po/w = -2.0-6.5
Discussion
The corona viral surface spike protein S is a type-I transmembrane glycoprotein that mediates initial host binding via the cell surface receptor angiotensin-converting enzyme 2 (ACE2), as well as the subsequent membrane fusion events required for cell entry. The SARSCoV surface spike protein S mediates viral entry into the host cell and includes two functional domains as follows: S1 (Gly13–Arg667) and S2 (Ser668–Thr1255). S1contains the host-specific receptor binding domain (RBD), whereas S2 mediates fusion between viral and host cell membranes (Xu, et al. [20]). Angiotensin-converting enzyme2 (ACE2) was identified as a functional receptor for the SARS-CoV (Li, et al. [21]). The recently determined structure of the S1-RBD in complex with the extracellular domain of ACE2 illustrates the structural basis for the initial step of virus-host recognition. As the mediator of host-specific SARS infection and a major viral surface antigen, the S protein is an attractive candidate for both vaccine development and immunotherapy. Marasco and co-workers in 2005 previously identified a potent neutralizing human monoclonal antibody against the S1 RBD, designated “80R,”fromtwonon-immune (i.e not restricted by B cell re-combination) human antibody libraries. 80R binds S1 with nano-molar affinity, blocks the binding of S1 to ACE2, prevents the formation of syncytia in-vitro (Sui et al. [22]), and inhibits viral replication in-vivo.
Deletion studies have shown that the 80R epitope on S1 is located in the minimal ACE2 binding domain, between residues 324 and 503 (Sui, et al. [23]). Terpenoid compounds used in this study were extracted from Buchholzia coriacea. Using Gas Chromatography-Mass Spectrophotometer (GC-MS), the compounds were quantified and characterized to give rutin, gallic acid, caffeic acid, ferulic acid, flavone, and phenol which have strong antioxidant properties. This study tends to predict the best inhibitor for SARS spike protein receptor binding domain (2GHV) from terpenoid compounds of Buchholzia coriacea using molecular docking approach. The binding affinity of the standard drug remdesivir was compared with other ligands of the compounds derived from B. coriacea. After docking of the phytochemicals with SARS spike protein receptor binding domain, the docking scores were as follows; rutin -7.545 kcal/mol, caffeic acid -7.297 kcal/ mol, flavone -7.276 kcal/mol, ferulic acid -6.696 kcal/mol, gallic acid -6.071 kcal/mol, phenol -4.732 kcal/mol and remdesivir -4.740 kcal/ mol [24-38]. From the result of ADME analysis, the blood brain barrier permeability of the tested compounds was nearly between the acceptable ranges which is very important for a drug to pass through those barriers rutin, gallic acid, caffeic acid, ferulic acid, flavone, and phenol showed QPlogBB value of -4.278, -1.661 -1.559, -1.066, 0.078, and 0.101 respectively.
Where the acceptable range is -3.0 to 1.2, rutin could not adhere to the QPlogBB result therefore, would not be able to penetrate blood brain barrier while the remaining compounds sailed through. The secondary metabolites: rutin, gallic acid, caffeic acid, ferulic acid, phenol and flavones showed the number of hydrogen bonds donor value of 9, 4, 3, 2, 1 and 0 respectively. Where acceptable range is 0.0-6.0 and also showed the number of hydrogen bonds acceptor value of 20.55, 4.25, 3.5, 3.5, 0.75 and 2.5 respectively. These are in the value of acceptable range (2.0-20.0) [39-59]. Rutin could not sail through the hydrogen bond donor and hydrogen bond acceptor because it has exceeded the acceptable range while phenol having the least number could also not sail through because it could not meet up the acceptable range. The other compounds were able to adhere to the range.
Rutin, caffeic acid, flavone, ferulic acid, gallic acid and phenol showed solvent accessible surface area value of 767.872, 389.661, 457.137, 408.488, 341.213 and 276.612 respectively where the acceptable range value is 300.0-1000. All compounds sailed through the solvent accessible surface area value except phenol which could not meet up the acceptable range. Predicted IC50 value for blocking Human ethera- go-go related gene (HERG) K+ channel for caffeic acid, ferulic acid, gallic acid and phenol were in the acceptable range (below -5), while rutin (-5.127) and flavone (-5.451) were above the acceptable range. The predicted octanol or water partition coefficient for the compounds were also analysed rutin, caffeic acid, flavone, ferulic acid, gallic acid and phenol showed -2.567, 0.539, 3.553, 1.389, -0.577 and 1.459 respectively where the acceptable range is -2.0-6.5. Human oral absorption rate was greater for flavone (100%), phenol (100%), ferulic acid (68.81%), caffeic acid (54.072%), gallic acid (41.457%) and rutin (0%) [60-70]. where the acceptable percentage < 25% is poor and > 80% is good. Caffeic acid, gallic acid and ferulic acid with the binding affinities -7.297 kcal/mol, -6.071 kcal/mol and -6.696 kcal/ mol greater than the binding affinity of the standard drug (remdesivir) -4.740 kcal/mol sailed through the 8 ADME analysis. Therefore, they could be lead compounds in the treatment of SARS-COV [71-74].
Conclusion
Conclusively, further wet research should be carried out on caffeic acid, gallic acid and ferulic acid using in-vivo analysis to validate the result. Clinical verification is needed to ascertain them as better drugs with little or no side effects in the management of SARS-COV.
We acknowledge the Director of Research, Center for Biosafety and Drug Development (CBDD), Adekunle Ajasin University, Akungba Akoko, Ondo State, Nigeria for allowing us to use his laboratory.
There was no conflict of interest declared by the authors of this manuscript.
The authors of this research work received no financial support.


