Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Anwaar S Chaudary1, Chenchang Jiang1, Yanglin Guo1 and Edward S Gasanoff1,2*
Received: May 15, 2024; Published: May 23, 2024
*Corresponding author: Edward S Gasanoff, STEM Research Center, Chaoyang Kaiwen Academy, 100018 Beijing, China and Belozersky Institute of Physico-Chemical Biology, M.V. Lomonosov Moscow State University, 119991 Moscow, Russia
DOI: 10.26717/BJSTR.2024.56.008895
This study describes the isolation of novel acidic and basic proteins from the venom of the Latrodectus pallidus (white widow spider), which exhibit high amino acid sequence homology to the 18.5 kDa isoforms of myelin basic proteins. This study explores the ability of model myelin membranes, composed of phospholipids and the acidic and basic proteins from spider venom, to absorb protons and form a non-bilayer lipid phase. The results of this study support a previously suggested concept by A.M. Morelli, which proposes that the myelin membrane may accumulate protons on its surface to store energy. The energy stored on membrane surface could then be used to drive proton circuits, potentially coupling hypothetical redox processes and ATP synthesis within the myelin membrane.
Spiders evolved over 300 million years ago, and through their long evolutionary history, spiders developed into animals with wellequipped venoms comprising a great variety of biochemical compounds designed to paralyze and/or kill their prey, rendering spiders the most successful venomous creatures in evolution [1]. Proteins constitute a major portion of spider venoms that display diverse pharmacological activities, representing vast evolutionary-edited natural pharmacopoeias that attract research efforts leading to the potential development of novel pharmaceuticals [2,3]. A few spider venom protein toxins have been used as invaluable ligands targeting various pharmacological targets. Some of these venom protein toxins are now undergoing preclinical and clinical studies for the treatment of multiple sclerosis, diabetes, and cardiovascular pathologies [4]. Spider venom proteins are also used in fundamental studies as crucial tools for probing electrophysiological processes in biological membranes [2]. For example, hanatoxins from Grammostola rosea and agatoxins from Agelenopsis aperta have been used as specific ligands of voltage-gated potassium and calcium channels [5,6]. In addition, spider venoms comprise a variety of stable small proteins with strong insecticidal activity that cause paralysis and/or lethality of insects by affecting ion channels, receptors, and enzymes [7]. This kind of research on the insecticidal activity of spider venom proteins holds high potential for the development of novel bioinsecticides, which are finding applications in green agricultural biotechnology [8,9].
The venoms of Latrodectus widow spiders, a genus of spiders in the family Theridiidae erected by Charles Athanase Walckenaer in 1805 [10], are probably the most studied spider venoms due to their extraordinary potent neurotoxins [11,12], which draw not only medical interest but also the general interest of scholars who use Latrodectus neurotoxins in studies of molecular mechanisms in neurobiology and pharmaceutical research [13]. The spiders of the Latrodectus genus comprise 32 species spread around the globe [14-16], with most of the species found in South and North Americas [10,17]. The most potent Latrodectus neurotoxin is α-latrotoxin, which targets the vertebrate central nervous system by attacking the presynaptic membrane of neurons, leading to increased intracellular Ca2+ concentration that triggers elevated exocytosis of neurotransmitters in the intermembrane synaptic cleft [13,18]. The less potent Latrodectus neurotoxins are α, β, γ, δ, ε-latroinsectotoxins, which bind to specific receptors on the presynaptic membrane of neuronal cells of insects, and α-latrocrustatoxin, which attacks the presynaptic membrane of crustaceans [18].
Overall, the neurotoxic activity of ‘latro-insecto-crusta-toxins’ is based on the binding of toxins to neuronal cells, which promotes the release of neurotransmitters either via stimulating exocytosis or through the formation of transmembrane tetrameric pores promoting specific Ca2+ permeability [18]. Noteworthy, it has been reported earlier that α-latroinsectotoxin from Latrodectus mactans venom promotes the conductivity of bilayer lipid membranes in the ion channel manner for divalent cations in the following order of decreasing permeability Ba2+ > Ca2+ > Mg2+ > Cd2+ > Zn2+ [19]. Besides the neurotoxins, Latrodectus venoms have been reported to contain metalloproteases, serine proteases, chitinases, hyaluronidases, venom allergen antigen 5-like proteins, etc. [13]. In addition, cationic proteins with antibacterial activity have been detected in Latrodectus geometricus venom [20]. In a recent study, the chromosome-level genome assembly of Latrodectus elegans spider venom with 55 identified toxin genes has been reported [21], which is more than the number of different types of venom proteins identified in all studied species of the Latrodectus genus. This finding suggests that there are still more unknown proteins to be identified in the venom of spiders of the Latrodectus genus.
The venom of Latrodectus pallidus, the white widow spider, which inhabits the deserts of Central Asia, is the least studied venom among species of the Latrodectus genus [22]. In 2014, the venom from Latrodectus pallidus was fractionated to reveal that, in addition to the types of proteins commonly found in venoms of the Latrodectus genus, such as α-latrotoxin, latrodectin, latroinsectotoxin, phospholipase D, hyaluronidase, and serine protease, there are two proteins, one acidic and another basic, both of 18.5 kDa, in the Latrodectus pallidus venom. These proteins have amino acid sequences which are not homologous to the amino acid sequences of proteins previously identified in venoms of the Latrodectus genus [22]. Interestingly, the amino acid sequences of the two proteins of 18.5 kDa from the Latrodectus pallidus venom are highly homologous to the sequences of myelin basic protein isoforms. In the present study, we used the Latrodectus pallidus venom acidic and basic proteins of 18.5 kDa as the protein component of the model myelin membrane, which included, as the lipid component, the major phospholipids of the myelin membrane, such as sphingomyelin, phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, and phosphatidylcholine, to examine the proton absorbing capacity and the polymorphic phospholipid transitions in the model myelin membrane. The results obtained in the present study support the innovative proposal that outlines the novel biological function of the myelin membrane related to the accumulation of protons on the myelin membrane surface to serve a function of proton capacitor, a novel concept suggested by Professor Alessandro M. Morelli [23]. The results of the present study also propose new tentative physiological roles for phospholipids and acidic and basic proteins of the myelin membrane.
Chromatography
The lyophilized venom from the Central Asian white widow spider Latrodectus pallidus was a gift from Prof. L. Ya. Yukelson (Institute of Biochemistry, Uzbekistan Academy of Sciences). The venom (200 mg) was dissolved in 1.5 ml of 5.0 mM Tris-HCl, pH 8.0 buffer and applied onto the cation-exchange CM Sephadex C-50 (Nanjing Duly Biotech Co. Ltd, Nanjing, China) borosilicate column (1.5 × 35 cm) equilibrated overnight in 5.0 mM Tris-HCl, pH 8.0 buffer at 25°C. The KCl gradient in the same buffer was administered as shown in Figure 1. Acidic protein was collected from the shaded area of F1 peak (Figure 1), then dialyzed against water in 1.0 kDa cutoff dialysis tube (Sigma Aldrich, Saint Louis, USA) and lyophilized on the vacuum freeze-drying lyophilizing machine YTLG-10 (Shanghai Yetuo Co., Ltd., China) at 2 Pa and –60°C for 12 hours. A sample from shaded area in F7 peak (Figure 1) was dialyzed and lyophilized as described above and then sub-fractionated by cation-exchange HPLC in a Syn- Chropak S300 column. A 20 mg/ml sample was dissolved in 0.02 M Bis-Tris, pH 7.3 and injected into a SynChropak S300 column and a linear gradient was established with 0.5 M HCl buffer 5 min after the sample was injected. Basic protein was collected after 17 min elution time as shown by shaded area in Figure 2. The basic protein was dialyzed and lyophilized as described above.
Electrophoresis
SDS-polyacrylamide gel electrophoresis was carried out on the Mini-PROTEAN Tetra Vertical Electrophoresis Cell using Mini-PROTEAN TGX precast 12% density gels (Bio-Rad Laboratories Co., Ltd., Shanghai, China). Protein samples were dissolved in 20mM Tris-HCl pH 7.0 buffer with 2 mM EDTA, 5% SDS, 10% DDT, 0.01% Bromophenol Blue, 2% glycerol at a final protein concentration 2 mg/ml and boiled for 10 min. A 15 μl portion of each protein sample and low molecular weight markers for SDS-PAGE (Thermo Fisher Scientific Inc., Shanghai, China) were applied into wells of the precast 12% density gel. The running buffer was 500 ml 1×TAE (Tris-acetate-EDTA) pH 7.0 including 7.2 g glycine and 0.2 g SDS. Electrophoresis proceeded for 50 minutes at 100 V. Isoelectric focusing (IEF) was carried out on the horizontal plate Isoelectric Focusing apparatus DYCP-37B (Beijing Liuyi Biotechnology Co., Ltd., China) using the Ready Gel Precast Gel with ampholyte of pH range 3–10.5 and the protein markers for IEF with pI range of 4.65–10.6 (Shanghai Yeyuan Biotechnology Co., Ltd., China). The pH gradient in the precast IEF gel was adjected by running electric power at 75 V for 30 minutes after which 5 μl of each protein sample (1mg/1ml H2O) and IEF protein markers were applied onto the precast IEF gel and the power of 90 V was run for 120 minutes. Amino acid residues sequencing: The detailed protocol for resolving the sequences of amino acid residues of acidic and basic proteins purified from the Central Asian white widow spider Latrodectus pallidus was described previously [22]. Briefly, the amino acid composition of proteins was determined with the High-Speed Amino Acid Analyzer LA8080 AminoSAAYA (Hitachi High-Tech, Shanghai Co., Ltd., China). Reduction and carboxymethylation of proteins were conducted as previously described [24]. Proteins were hydrolyzed for 5 hours at 37°C with trypsin at 1:50 w/w protein to trypsin ratio. The carboxymethylated protein hydrolysate was chromatographed on Chromo- Beads resin (Koram Biotech Corp., Seul, Korea) and then further purified by paper electrophoresis and chromatography. The amino acid sequence of the protein fragments was determined as described previously [25]. The carboxymethylated proteins were incubated for 20 hours at 25°C in 70% formic acid. The cyanogen bromide cleavage was done using a 100-fold excess of reagent. The products of the carboxymethylation and cyanogen bromide cleavage were purified on Sephadex G-25 in 0.1 M NH4–HCO3 buffer. Paper chromatography was used to isolate the chymotryptic peptides of the cyanogen bromide fragment of proteins.
The N-terminal sequences of carboxymethylated proteins and the cyanogen bromide fragment forming the C-terminus of the original proteins were resolved on a Beckman model 890C sequencer (Beckman Instruments Inc., Fullerton, CA, USA) and the C-terminus sequence was resolved by means of carboxypeptidase A. Total lipid extract isolation and purification: Wistar rat liver was homogenized in a Warning Blender 700G for 3 minutes in 300 ml chloroform and 200 ml methanol, then100 ml of methanol and 200 ml dd-H2O was added and the mixture was blended for another 5 minutes. Homogenate was centrifuged at 200 g for 20 minutes and aqueous layer of methanol and dd-H2O was discarded. The chloroform phase with lipids was washed with 300 ml saline and centrifuged at 200 g for 15 minutes after which saline layer was discarded, and the chloroform phase was washed two more times. The lipids in chloroform were filtrated through the Whatman No.1 filter paper and the lipids were washed from the filter paper with 30 ml methanol.
Preparation of phospholipid specific polyvalent antibodies: Polyvalent antibodies specific for the selected phospholipids were developed using adult male Wistar rats. The adjuvant-primed animals received injections of the synthetic polar heads of phospholipids: one animal was injected with one type of phospholipid polar head. The synthetic polar heads of phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol, phosphatidylserine and sphingomyelin were from NOF America Co., White Plains, USA. Ascites fluids were collected from the peritoneal cavity of animals two weeks after injection. The polyvalent antibodies from ascites fluids were purified on a Protein-A-Sepharose column (Pharmacia, Uppsala, Sweden). Purification of individual phospholipids by immunoaffinity chromatography: The borosilicate columns for immunoaffinity chromatography were loaded with the Sephadex G-25 resin containing covalently linked antibodies specific to the polar heads of either phosphatidylserine, sphingomyelin, phosphatidylinositol, phosphatidylcholine or phosphatidylethanolamine with each column containing only one type of antibodies, so overall five different immunoaffinity columns were prepared.
The antibodies were covalently linked to the Sephadex G-25 resin using Pierce™ Traut’s Reagent 2-iminothiolane Kit and N-succinimidyl 3-(2-pyridyldithio)propionate Kit (Thermo Fisher Scientific Inc., Shanghai, China) according to the manufacture’s protocol. The Sephadex G-25 resin with linked antibodies was equilibrated in 1.0 mM Tris-HCl, pH 7.0 buffer at 25°C for three hours. A 5 ml portion of the concentrated total lipid extract in methanol was applied into each of five immunoaffinity columns and the individual phospholipid fractions eluted from the columns were pulled into tubes with 15 ml chloroform and centrifuged at 200 g for 15 minutes to discard aqueous layer. Chloroform from the tubes was removed by drying with vacuum pump for one hour. The mass of phospholipid obtained was determined as the different in mass between the tube with dry phospholipid and the same empty tube. The concentration of stock solutions of phospholipids in chloroform of 0.1 M was prepared assuming molecular masses of phospholipids: 716 g/mol for phosphatidylethanolamine, 887 g/mol for phosphatidylinositol, 768 g/mol for phosphatidylcholine, 792 g/mol for phosphatidylserine and 734 g/ mol for sphingomyelin.
Preparation of unilamellar liposomes: The unilamellar phospholipid liposomes for the studies of ability of liposomal membrane to absorb protons, that is to serve as protons capacitor, were prepared by the treatment of aqueous lipid dispersions with ultrasonic wavelength frequency. Aliquots of phospholipids in chloroform containing molar ratio 4 to1 of phosphatidylcholine and either phosphatidylethanolamine, phosphatidylserine, sphingomyelin or phosphatidylinositol were dried with vacuum pump for 5 hours at 25°C for complete removal of chloroform, that is until the chloroform free phospholipid films were formed. Phospholipid films were then hydrated with dd- H2O pH 7.0 at phospholipid concentration 10–5 M and the resulted phospholipid dispersion was sonicated with the Ultrasonic Dispenser Yt-JY96-IIN (Shanghai Yetuo Technology Co., Ltd., China) at 22 kHz in helium atmosphere for 15 minutes at 4°C. Liposomes were then incubated for 15 hours at 15°C. The unilamellar phospholipid liposomes modified by acidic and basic proteins were prepared by adding equal molar amounts of acidic and basic proteins into the liposome sample at a total protein concentration 2×10–7 M.
To prevent electrostatic interaction of acidic and basic proteins in solution, the acidic protein was added first to liposome samples and incubated for 15 minutes with the continuous stirring of liposome solution with the magnetic stirrer JB-3 (Shanghai INESA Scientific Instruments Co. Ltd., China). Then the basic protein was added to the liposomes which were already modified by acidic protein and liposomes were incubated for another 15 minutes. The pH readings were taken in pure dd-H2O, in liposome samples and in liposome samples modified by the acidic and basic proteins. For the sake of uniform pH readings, all liposome samples treated or untreated with the proteins were continuously stirred with the magnetic stirrer. The proton concentration in pure dd-H2O and liposome samples treated or untreated with the proteins were determined from the measured pH values by using the following conversion: [H+] = 10–pH. 1H-NMR studies of unilamellar liposomes: For the 1H-NMR studies 2H2O was used instead of dd-H2O to prepare sonicated unilamellar liposomes according to protocol described above. Phospholipid concentration in liposomes for the 1H-NMR studies was 1.4 × 10–2 M and protein concentration was 1.75 × 10–4 M. Liposomes were treated with 10 μl of saturated K3Fe(CN)6 solution per 1 ml of liposomes. 1H-NMR spectra from N+(CH3)3 groups of unilamellar liposomes were recorded using a Varian XL-200 spectrometer (USA) at an operating frequency of 200 MHz at 25°C. The width of the 90° pulse was 8.7 μs, the relaxation delay was 50 μs and the acquisition time for free induction signal was 1 s. The integral intensity of 1H-NMR signals from choline groups was measured in triplicate and variation among triplicate readings was never higher than 6%.
Statistics: For each data point coming from the pH readings of dd-H2O and the liposome samples, they were prepared in triplicate, and the standard deviation between the readings was within 0.09% of the means. The pH readings coming from the four types of liposome samples and from the dd-H2O sample were statistically compared by using an ANOVA test. For statistical comparison of pH readings between liposome samples and liposome samples treated with proteins, the T-test was used. The p-values less than 0.05 were considered as statistically significant differences.
We fractionated the Central Asian spider Latrodectus pallidus venom by the cation-exchange column chromatography to purify acidic and basic proteins. We used one column to purify acidic protein and two columns to purify basic protein. The chromatograms derived from the two column fractionations are shown in Figure 1. Acidic protein was collected from the shaded area of the F1 peak in the chromatogram obtained by fractionation on the CM Sephadex C-50 column (Figure 1A). To isolate the basic protein, a fraction of proteins was collected from the shaded area of the F7 peak (Figure 1A), and this fraction of proteins was then sub-fractionated by cation- exchange HPLC in a SynChropak S300 column. The basic protein was isolated from the shaded area of the fraction shown in Figure 1B. Both the acidic and basic proteins were homogeneous by SDS-PAGE and isoelectric focusing electrophoresis (Figure 2). The molecular mass of both proteins was somewhat below 21.1 kDa, and the pI values were around 4.65 and slightly above 10.6 for the acidic and basic proteins, respectively. The amino acid sequences of the acidic and basic proteins, which were resolved according to the protocol described in the Materials and Methods, are aligned in Figure 3.
Amino acid sequences of both proteins are highly homologous to the human myelin basic protein isoforms [26]. In addition, both acidic and basic proteins show 100% sequence homology to the myelin basic protein isoforms from various species in the highly conserved amino acid sequence regions [27] (Figure 3). The calculated molecular mass and the isoelectric points of the Latrodectus pallidus venom acidic and basic proteins match exactly the most abundant isoforms in the adult human myelin with molecular mass 18.5 kDa [26]. These findings prompted us to use acidic and basic proteins in the studies of electrophysiological and structural properties of model myelin membrane. We have incorporated acidic and basic proteins from the Latrodectus pallidus venom into the model myelin membrane to investigate whether the model myelin membrane can absorb protons on its surface from the bulk water and undergo polymorphic phospholipid transitions.
The myelin membrane in the vertebrate central nervous system is mostly made of five phospholipids: phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), and sphingomyelin (SM) [28-31]. In terms of the protein component of the membrane, the isoforms of myelin basic protein are one of the most abundant components of the myelin membrane, the primary role of which is to maintain a compact myelin sheath [32]. PC is the dominant phospholipid that stabilizes the bilayer structure in most, if not all, biological membranes, including the myelin membrane [32-34]. The role of the other four phospholipids is not clear, but it has been suggested that each of those phospholipids plays a particular purpose in the functional activities of the myelin membrane [32]. In the present work, for the studies of proton absorption by the membrane surface, we have prepared four types of model myelin membranes, each of which includes PC and one of the four phospholipids of a particular function with the molar percentage ratio of 80% for PC and 20% for either PE, PS, PI, or SM, with a total phospholipid concentration of 10–5 M.
For the protein component of the four types of model myelin membrane, the total protein concentration of acidic and basic proteins of an equal molar ratio was 2×10–7 M. For the studies of polymorphic transitions of phospholipids in the model myelin membranes, we prepared the same four types of myelin membranes with the molar percentage ratio of 80% for PC and 20% for one of the phospholipids of a functional group, but we used different total phospholipid and protein concentrations, which were 1.4 × 10–2 M for phospholipids and 1.75 × 10–4 M for proteins. The model membrane system for myelin membranes was unilamellar liposomes, both for the studies of proton absorption by the membrane surface and for the studies of polymorphic phospholipid transitions. To assess the absorbance of protons by the myelin membrane liposomes, we measured the difference between pH values in pure dd-H2O and in dd-H2O solutions of liposomes from four models of myelin membranes, which did not include acidic and basic proteins (Table 1).
Table 1: The pH readings taken in triplicate at 25°C in dd-H2O and in dd-H2O solutions of liposomes made of either phosphatidylcholine (PC) + phosphatidylserine (PS), phosphatidylcholine + phosphatidylinositol (PI), phosphatidylcholine + phosphatidylethanolamine (PE) or phosphatidylcholine + sphingomyelin (SM). The mean values of pH readings, standard deviations (SD) and the ANOVA p-value are also given in the Table 1. The concentration of phospholipids in all samples is 10–5 M.
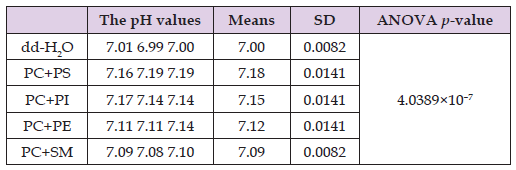
Preparations of liposomes in dd-H2O produced basic pH values (pH above 7.0), which is a result of protein absorption by the surface of the liposomal membrane. As shown in Table 1, the PS-containing membrane (PC+PS) demonstrates the highest change in pH towards basic values (7.18), indicating that the PC+PS membrane is the strongest proton capacitor among the four models of myelin membranes. This is due to the two acidic moieties, carboxyl and phosphate groups, in the PS polar head (Figure 4). Although the other three model myelin membranes each contain one of the ‘functionally-driven’ phospholipids (PI, PE, or SM), all of which have only one acidic moiety (phosphate group) in their polar heads, their proton absorption abilities differ and decrease in the following order: PI – pH 7.15, PE – pH 7.12, and SM – pH 7.09. The highest ability of PI to absorb protons among the three ‘functional’ phospholipids can be explained by the polar hydroxyl groups of the inositol moiety in the polar head of PI (Figure 4), which have the potential to absorb protons via the formation of coordinate bonds with the lone pair of electrons on the oxygen atoms of the hydroxyl groups.
The higher ability of PE to absorb protons compared to SM can be attributed to the larger size of the choline group (–N+(CH3)3) in SM versus the amino group (–N+H3) in PE (Figure 4). The smaller size of the amino group allows better access for protons to the phosphate group of PE than the choline group of SM. It should be noted that the ANOVA p-value for the statistical differences in pH values is very small within the group including pure dd-H2O and dd-H2O solutions of the four models of myelin liposomal membranes, indicating that the differences are statistically significant. The effect of the equimolar mixture of acidic and basic proteins (Pr) incorporated into the four models of myelin liposomal membranes is shown in Table 2. In all four membrane models, the incorporation of Pr resulted in a further increase in pH values, demonstrating that Pr contributes to the proton absorption of the myelin membrane. Thus, Pr enhances the myelin membrane’s ability to act as a proton capacitor. The largest increase in pH value (1.05) triggered by Pr was observed in the PS-containing membrane. The second largest increase in pH value (1.03) was observed in PE-containing membranes.
Table 2: Comparison of pH values in samples of liposomes made of either phosphatidylcholine (PC) + phosphatidylserine (PS), phosphatidylcholine + phosphatidylinositol (PI), phosphatidylcholine + phosphatidylethanolamine (PE) or phosphatidylcholine + sphingomyelin (SM) and the same liposomes treated with a mixture of acidic and basic proteins (Pr). The pH readings mean values, standard deviations (SD) and the T test p-values are also given in the Table 2. The concentrations of phospholipids and Pr are 10–5 M and 2×10–7 M respectively.
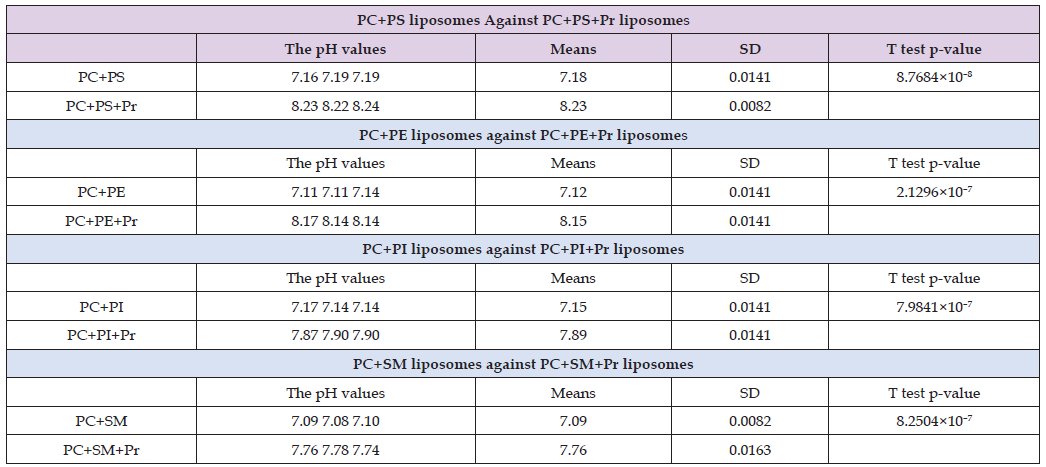
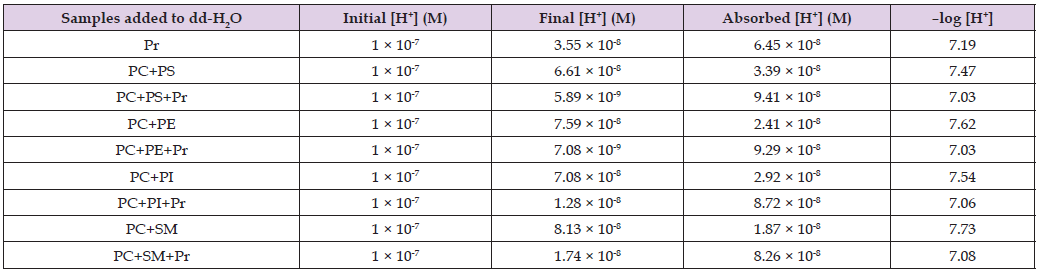
The increase in pH value triggered by Pr in PC+PI and PC+SM was significantly lower, at 0.74 and 0.67, respectively. The very low T-test p-values comparing the statistical differences between pH values in model myelin membranes not containing Pr and those containing Pr confirm that the increases in pH values triggered by Pr are statistically significant. To better present the differences in proton capacitor power between the four models of myelin membranes in the absence and presence of Pr, the concentrations of absorbed protons are shown in Figure 4 as the negative logarithm of [H+] absorbed by the model myelin membranes. It should be noted that the lower the negative logarithm of [H+] value, the higher the concentration of H+ ions absorbed. Thus, by comparing the heights of the bars in Figure 4, one can conclude that the myelin membranes containing Pr exhibit higher proton capacitor power than myelin membranes without Pr. Additionally, one can conclude that among the myelin membranes containing Pr, the PC+PS and PC+PE membranes have a higher proton absorbing capacity than the PC+PI and PC+SM membranes (Table 3).
Table 3: The concentration of H+ ions in dd-H2O (initial H+ concentration), concentration of H+ ions after adition of proteins, liposomes or liposomes modified with proteins to dd-H2O (final H+ concetration) and concentration of H+ ions absorbed by proteins, liposomes or liposomes modified with proteins (absorbed H+ concentration). Concentrations of phospholipids and proteins in liposome samples are 10–5 M and 2 × 10-7 M respectively. Pr – mixture of acidic and basic proteins, PC – phosphatidylcholine, PS – phosphatidylserine, PE – phosphatidylethanolamine, PI – phosphatidylinositol, SM – sphingomyelin. Column –log [H+] shows negative logarithm values of absorbed [H+].
To study the permeability and polymorphic transitions of phospholipids in the four models of myelin membranes in the absence and presence of Pr, we used 1H-NMR spectroscopy of unilamellar liposomes in the presence of K₃[Fe(CN)₆]. The methodology and advantages of unilamellar liposome 1H-NMR spectroscopy with paramagnetic Fe(CN)₆³⁻ ions are described in detail in our previous publications [35-38]. Briefly, Fe(CN)₆³⁻ ions interact with the choline (N+(CH₃)₃) groups of the outer monolayer of the liposomal membrane, shifting the choline signal of the outer monolayer to higher values of the applied magnetic field, while the signal of the choline groups from the inner monolayer remains unchanged. This results in the splitting of 1H-NMR signals from the outer and inner monolayers of the liposomal membrane. Should the liposomal membrane become permeable to Fe(CN)₆³⁻ ions, which would allow interaction with the choline groups of the inner membrane, the signal of the choline groups from the inner monolayer would shift to the field of the outer monolayers, causing the inner monolayer signal to visually disappear.
As shown in Figure 5, all four model myelin liposomal membranes in the absence of Pr were not permeable to Fe(CN)₆³⁻ ions. The higher intensity of the 1H-NMR signals in the PC+SM liposomes compared to the other three types of liposomes is due to both PC and SM having a choline group in their polar heads, while in the other three types of liposomes, only PC has a choline group. The addition of Pr to the liposome samples resulted in the broadening of 1H-NMR signals due to the restriction of molecular mobility of lipids by Pr. However, the addition of Pr did not make the membrane in any of the four liposome samples permeable to Fe(CN)₆³⁻ ions, as the signals from the inner monolayers remained intact. Notably, in the PC+PS+Pr and PC+PE+Pr liposomes, a small signal appeared in the higher field from the signal of the outer monolayer (Figure 5). This signal originated from the formation of a non-bilayer lipid phase [35-38] and was more pronounced in the PC+PE+Pr membrane. The formation of the non-bilayer lipid phase in model membranes containing PS and PE, triggered by interaction with basic proteins, was observed previously [35,38-41]. It is evident that the non-bilayer lipid phase in model myelin membranes containing PS or PE was induced by the basic protein from the white widow spider venom (Figures 5 & 6).
It has been suggested that physiologically active proteins in animal venom have evolved by mimicking the structure and functions of body proteins with important functional activity [42]. For example, many phospholipases isolated from snake and insect venoms resemble structurally and functionally the phospholipases that regulate inflammatory processes and lipid metabolism in biological membranes [42,43]. Rattlesnake venom metalloproteinases have been reported to act similarly to body metalloproteinases involved in controlling the homeostasis of blood and intercellular fluids [42,44]. Cationic proteins isolated from cobra venom have been shown to phenocopy the membranotropic activities of the C-8 protein from the Fo sector of ATP synthase [45-47]. In this research study, we present acidic and basic proteins isolated from the venom of the white widow spider Latrodectus pallidus, with amino acid sequences highly homologous to the isoforms of myelin basic proteins of the vertebrate central nervous system [25,26]. It is therefore reasonable to assume that the physiological activities of these acidic and basic proteins from Latrodectus pallidus venom resemble those of the myelin basic proteins of the vertebrate central nervous system.
Myelin basic proteins exist as isoforms that differ in size and charge [48]. The size isoforms are produced by alternative splicing of an mRNA transcript [49], while the charge isoforms result from post-translational modifications that decrease the basic charge, affecting the functional activities of the myelin basic proteins [26]. The size isoforms of human myelin basic proteins include proteins of 17.2, 18.5, 20.2, and 21.5 kDa, with the 18.5 kDa isoform being the most abundant and the most studied [26,49]. There are eight charge isoforms of the 18.5 kDa myelin basic protein, termed C1–C8. The C1 isoform has the highest positive charge and is the least modified after post-translational modification [26]. The charge isoforms C2–C6 are modified by phosphorylation, deamidation, and deamination, while C8 is predominantly modified by peptidyl arginine deiminase, which converts positively charged arginine to neutral citrulline. This conversion can involve up to 11 arginine residues, leading to an overall loss of positive charge of +11 [50]. The isoelectric points of the 18.5 kDa isoforms range from 4.5 for C8 to 11 for C1 [26], which is very similar to the isoelectric points of acidic and basic proteins of 18.5 kDa from the venom of the Latrodectus pallidus spider. This suggests that acidic and basic proteins from spider venom may phenocopy the functions of the C1 and C8 isoforms of the 18.5 kDa myelin basic protein. The myelin sheath is a repetitive multilayer of tightly packed myelin membrane bilayers held together by proteins in myelin membranes [26]. The most long-lived proteins in the body are myelin basic proteins [51,52]. This suggests the importance of myelin basic proteins in the stability of the supramolecular structure of the myelin sheath [26]. It is believed that the C1 isoform functions by tightly attaching to acidic phospholipids of myelin bilayer membranes, stabilizing the tight packing of the myelin sheath [26]. Overall, C1, C2, and C3 are most likely responsible for myelin stability [26], while C8, which is abundant in childhood and scarce in adults, seems to play a role in development and is important in the formation of myelin rather than its stability [50]. However, the exact molecular mechanism of the functional role of C8 is not understood. Notably, the pathogenesis of multiple sclerosis (MS) is linked to abnormal changes in the 18.5 kDa isoform composition caused by an abnormal increase in the activity of peptidyl arginine deiminases, leading to the loosening of the tight packing of the myelin sheath [26,53-56].
The common explanation of the role of the myelin sheath is that it serves as an insulator of neuronal axons to promote rapid and saltatory conduction of nerve impulses [26,57]. However, nerve impulses are not transmitted through nerve fibers in the same way electrons move through a metal wire [57]. It is not clear how myelin insulates neurotransmission or what the insulation of nerve impulses means. It is obvious that even if myelin serves as an insulator in some way, it is far more than just an insulator [57]. A breakthrough discovery revealed aerobic ATP synthesis in the myelin sheath, which was unexpected as myelin is devoid of mitochondria [58-61]. However, oligomycin titration experiments have determined the presence of F1 subunits of ATP synthase in myelin [59], and the presence of respiratory complexes in lipid rafts of myelin was also established [62,63]. These discoveries suggest a way for delivering mitochondrial respiratory components to the lipid rafts in myelin.
Professor Alessandro Morelli of Genoa University proposed that mitochondria, which have their own DNA for respiratory complexes and ATP synthase to drive oxidative phosphorylation, deliver all the protein complexes necessary for ATP synthesis to the myelin sheath with the help of the endoplasmic reticulum (ER) [64], with which mitochondria are closely associated in cytoplasmic space and function [65,66]. It has been determined that mitochondria produce necessary vesicles providing the ER with the necessary oxidative phosphorylation complexes [67,68]. Aerobic ATP synthesis has also been observed in structures devoid of mitochondria such as rod outer segment discs [69-72], platelets [73], cell plasma membranes [74-78], exosomes, and microvesicles [79], strongly implying that the oxidative phosphorylation machinery is transferred to these extra-mitochondrial sites via the ER [80].
It has been shown by freeze-fracture X-ray crystallography that particles of 8.6 nm, the dehydrated F1 subunits of ATP synthases, are exposed on both sides of the myelin sheath [81,82]. This bi-faced orientation of F1 subunits in the myelin sheath does not agree with Mitchell’s concept of delocalized proton coupling, where the gradient of proton concentrations in bulk water across the inner mitochondrial membrane drives the proton movement through ATP synthases with the F1 subunits exposed only on the matrix side. However, the orientation of F1 subunits on both sides of the myelin sheath agrees with the concept of localized coupling, where protons are absorbed and move along the surface on both sides of the myelin membrane, establishing a proton circuit built entirely inside a major dense line of myelin [80]. Professor Morelli suggests that extra-mitochondrial synthesis of ATP can take place on any single membrane as long as the membrane surface can localize (absorb) protons to act as a ‘proton capacitor’, so the accumulated positive charge can support a proton circuit in which protons move along the membrane surface by the Grotthuss mechanism [83].
From the respiratory complexes to the F1 subunit of ATP synthase, and in the middle of the F1 subunit, protons turn to the hydrophobic center of the membrane through which protons move back to the respiratory complexes [23]. Thus, the proton circuit, taking place entirely inside a single membrane, couples respiration with ATP synthesis [23,64,80]. In our view, the experimental evidence described above supports localized proton coupling, which is based on the membrane’s ability to absorb protons on its surface. However, we note that protons cannot move as charge through the low dielectric environment in the center of the membrane, and there must be a vehicle to transfer protons through the hydrophobic environment to the respiratory complexes.
The experimental results of our study on model myelin membranes with acidic and basic spider proteins, acting as isoforms of myelin basic proteins, showed that these membranes absorb protons on the membrane surface.
The protons are absorbed by both the phospholipid polar heads and the spider proteins, primarily by acidic proteins rather than basic ones. The ability of phospholipids to absorb protons differs depending on the physical-chemical properties of the phospholipid polar head. Additionally, we observed the formation of a non-bilayer phase in model myelin membranes made of PC+PS+Pr and PC+PE+Pr. We have previously observed the formation of a non-bilayer phase through the interaction of PS with basic proteins. The non-bilayer propensity of PE was also described previously, and it is strongly enhanced by interaction with basic proteins. Therefore, we can conclude that the non-bilayer phase in model myelin membranes was triggered by the interaction of spider basic protein, but not by spider acidic protein, with PS and PE. It should be noted that the highest ability to absorb protons was observed in PC+PS+Pr and PC+PE+Pr membranes, which contain the non-bilayer phase. In our previous studies, we have demonstrated that the non-bilayer phase in the inner mitochondrial membrane, triggered by basic proteins, stabilizes the intermembrane association between adjacent crista membranes, facilitating higher activity of ATP synthase [35,46,84,85].
One of the mechanisms for enhancing ATP synthase activity facilitated by the non-bilayer phase was recently proposed by us, in which inverted micelles transfer protons across the hydrophobic environment of the crista membrane in the inner aqueous volume of inverted micelles [35,86]. The non-bilayer phase observed in our study in model myelin membranes likely exists in the form of inverted micelles, which transport protons in the myelin membrane’s hydrophobic environment, not across the membrane but through the membrane center from the F1 subunit to the respiratory complexes to complete the proton circuit inside a single membrane, as proposed by Professor Morelli [64,80].
The experimental results of our study support the new concept of the physiological role of the myelin sheath, related to the accumulation of energy through membrane action as a proton capacitor, as proposed by Professor A.M. Morelli. The stored energy is utilized for the production of ATP synthase through localized proton circuits that couple respiratory complexes with ATP synthases. According to Professor A.M. Morelli’s proposal, the generated ATP molecules in the myelin sheath could be used by ionic pumps in neuronal axons for the rapid transmission of nerve impulses. Our results suggest that the acidic isoforms, but not the basic isoforms, of myelin basic proteins, along with phospholipid polar heads, are responsible for the absorption of protons on the membrane surface. Additionally, our results indicate that basic isoforms of myelin proteins interact with acidic PS and neutral PE to promote the formation of a non-bilayer phase. The non-bilayer phase may promote the tight association of adjacent myelin membranes and facilitate the transport of protons through the hydrophobic environment in the center of the myelin membrane to couple respiratory complexes with ATP synthase.
The results obtained in this study warrant further investigation of acidic and basic proteins from the Latrodectus pallidus spider venom in model myelin membrane systems. This research could lead to a better understanding of the molecular mechanisms of neurotransmission and neurodegeneration, as well as the potential development of novel pharmaceutical products that may alleviate or halt the process of myelin sheath degradation in aging and diseases.
Dr. Anwaar S Chaudary is grateful to Prof. L. Ya. Yukelson of the Institute of Biochemistry, Uzbekistan Academy of Sciences for Latrodectus pallidus spider venom given as a gift. The research project was supported by the start-up grant from the Beijing Chaoyang Kaiwen Academy to the STEM Research Center.


