Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Akshay Suresh Patil and Yan Xu*
Received: February 15, 2024; Published: March 11, 2024
*Corresponding author: Yan Xu, Department of Chemistry, Cleveland State University, Cleveland, USA
DOI: 10.26717/BJSTR.2024.55.008706
Allergic rhinitis, an immune disorder affecting over 80 million people in the United States annually, is characterized by the immune system’s excessive reaction to external allergens. Common synthetic treatments for this condition are often associated with side effects, including dizziness, weakness, and an increased heart rate. This has led to a growing interest in herbal remedies, which typically exhibit fewer adverse effects. Among these, Astragalus radix (AR), a staple in traditional Chinese medicine known for its immunomodulating and anti-inflammatory properties and used in treating allergies for centuries, stands out. Our study delves into the components of AR, targeting the proteins involved in allergic rhinitis. We utilized various databases to collect data on AR components, their protein targets, and allergic rhinitis disease genes. A protein-protein interaction network was constructed, facilitating the enrichment of biological functions (processes and pathways). The proteins associated with the enriched functions relevant to allergic rhinitis (p < 0.05) were used to construct a target-target-interaction (TTI) network. We employed modularity analysis and an integrated centrality scoring approach on this network, identifying four proteins (ALOX5, IL2, TNFα, and TLR4) as pivotal targets for allergic rhinitis. Further, we evaluated the interaction between these pivotal proteins and AR’s components through molecular docking simulations. Our findings highlight four bioactive components of AR - kaempferol, daidzein, calycosin 7-O-glucoside, and astragaloside III - each exhibiting the lowest binding energies, suggesting their potential as antagonists to regulate these pivotal proteins in allergic rhinitis management.
Keywords: Allergic Rhinitis; Astragalus Radix; ALOX5; IL2; TNFα; TLR4; Kaempferol; Daidzein; Calycosin 7-OGlucoside; Astragaloside III
Abbreviations: ALOX5: Lipoxygenase-5; AR: Astragalus Radix; BC: Betweenness Centrality; CC: Closeness Centrality ; DC: Degree Centrality; DCdb: Drug Central Database; DGIdb: Drug-Gene Interaction Database; DL : Drug Likeness; EC: Eigenvector centrality; ETCM : The Encyclopedia of Traditional Chinese Medicine; GDA : Gene Disease Association; GO: Gene Ontology; IC: Integrated Centrality; IL2 : Interleukin 2; KEGG: Kyoto Encyclopedia of Genes and Genomes; Max: Maximum; Min: Minimum; OB: Oral bioavailability; PPI: Protein-Protein Interaction; SP: Super Pred; STP: Swiss Target Prediction; TCM: Traditional Chinese Medicine; TCMID : Traditional Chinese Medicine Information Database; TCMSP: Traditional Chinese Medicine Systems Pharmacology; TLR4: Toll-Like Receptor 4; TNFα: Tumor Necrosis Factor Alpha; TTD: Therapeutic Target database; TTI: Target-Target interaction
Allergic rhinitis, commonly referred to as seasonal allergies or hay fever, is a widespread immune system disorder. This condition, characterized by an overactive immune response to external particles, affects approximately 80 million individuals annually in the United States alone (Ng, et al. [1,2]). Allergic rhinitis specifically denotes nasal inflammation triggered by allergens like pollen, hay, mold spores, or dust. While bacteria are not a primary cause, they can contribute to allergic rhinitis through infections in congested nasal passages. The immune response involves dendritic cells capturing and processing allergens, activating T cells, which then prompts B cells to produce IgE antibodies attaching to the FCε receptor. The subsequent binding of allergens to IgE causes the crosslinking of IgE-FCε complexes, releasing histamines, leukotrienes, and other inflammatory mediators through degranulation (Bernstein, et al. [3,4]). The uncontrolled release of inflammatory cytokines, interleukins, interferons, and chemokines causes allergic reactions, leading to sneezing, runny nose, itching, watery eyes, and swelling (Kay [5]). Pathways like Toll-like receptor signaling, FCε-RI signaling, histamine metabolism and degradation, and arachidonic acid metabolism play crucial roles in allergic rhinitis development (Ahmad, et al. [6-9]). The current treatment options for allergic rhinitis include anti-allergic medications such as antihistamines (levocetirizine), intranasal corticosteroids (fluticasone), decongestants (pseudoephedrine), bronchodilators (ipratropium), anti-inflammatory agents or mast-cells stabilizers (cromolyn), which are used to prevent or reduce allergic reactions (Bernstein, et al. [3,10]).
In cases where the reaction worsens, artificial epinephrine shots are administered. These medications work by dilating the constricted nasal passages, actively inhibiting the production of inflammatory signaling molecules, or blocking the receptors to initiate the inflammatory cascade. However, most of these medications are synthetic and often lead to side effects, such as drowsiness, dizziness, headache, rapid heart rate, muscle weakness, etc. Long-term use of these medications may also negatively impact the immune system (Bender [11]). Consequently, plant-based supplements are gaining popularity as a safer alternative. Astragalus radix (AR), or Huang Qi, is derived from the dried roots of the Astragalus membranaceus tree and is extensively used in traditional Chinese medicine (TCM) due to its potent immunomodulatory effects, either alone or in combination with other herbs (Fu, et al. [12]). AR is a key ingredient of many TCM formulas, such as Yu Ping Feng decoction (Zuo, et al. [13]), Huangqi Sijunzi decoction (Zhang, et al. [14]), Huang Qi Jian Zhong Tang (Nöst, et al. [15]), etc., and is known for enhancing bodily resistance to allergens and boosting vitality. It is widely recognized as a valuable herbal medicine that increases energy levels and effectively balances the immune system (Z Chen, et al. [16]). The immunomodulatory and anti-inflammatory properties of AR were reported to regulate nitric oxide, cytokines, and interleukins (Choi, et al. [17,18]). Other studies also showed that AR promotes wound healing and has hepatoprotection, antioxidant, anti- hyperglycemic, and anti-viral properties (Fu et, al. [12,19]). The use of AR for weakness, anemia, fever, fatigue, and loss of appetite was also reported (Yu, et al. [19]).
Despite its wide application, the specific components of AR responsible for its therapeutic effects remain under-researched. According to the TCM database, AR contains nearly 90 components, primarily flavonoids, saponins, and polysaccharides. Some of these components exhibit pharmacological activity and therapeutic efficacy by interacting with various disease genes in biological processes and pathways (Fu, et al. [12]). Network pharmacology offers a comprehensive approach to uncovering these components by analyzing bioinformatics and cheminformatics data (Hopkins, et al. [20,21]). This method aids in identifying bioactive compounds in AR that could be potential therapeutic agents against allergic rhinitis. Additionally, molecular docking simulation, an in-silico validation method, helps understand the interactions (i.e., binding packets and energies) between these components and their protein targets (Agarwal, et al. [22,23]). The present study aims to identify the key bioactive components of AR and their pivotal protein targets for the treatment of allergic rhinitis and gain an understanding of AR’s molecular action mechanisms.
AR Component Acquisition
The components examined for AR were gathered and integrated from several databases, including Traditional Chinese Medicine Systems Pharmacology (TCMSP: https://old.tcmsp-e.com/index.php) (Ru, et al. [24]), Traditional Chinese Medicine Information database (TCM-ID: http://bidd.group/TCMID/index.html) (X Chen, et al. [25]), and The Encyclopedia of Traditional Chinese Medicine (ETCM: http://www.tcmip.cn/ETCM/) (HY Xu, et al. [26]). These databases provide comprehensive information on the physicochemical properties and drug-likeness of the herb's components, such as molecular weight, A (log p), oral bioavailability (OB), drug-likeness (DL), H-donor, H-acceptor, and blood-brain barrier values. The selection criteria for screening the AR candidate components in this study were based on OB and DL, as the primary route of administration for AR is via oral ingestion.
Protein Target Acquisition
Protein targets for both AR components and allergic rhinitis were obtained through web servers. Specifically, protein targets for screened AR components were acquired using Similarity Ensemble Approach (SEA: https://sea.bkslab.org/) (Keiser, et al. [27,28]), Swiss Target Prediction (STP: http://www.swisstargetprediction.ch/) (Daina, et al. [29]), Super Pred (SP: https://prediction.charite.de/) (Nickel, et al. [30]), Herb Ingredients Targets platform (HIT 2.0: http://hit2.badd-cao.net/) (Yan, et al. [31]), Therapeutic Targets Database (TTD: https://db.idrblab.net/ttd/) (Zhou, et al. [32]), Drug Central Database (DCdb: https://drugcentral.org/) (Ursu, et al. [33]), Drug-Gene Interaction Database (DGIdb: https://www.dgidb.org/) (Freshour, et al. [34]), and BindingDB (https://www.bindingdb.org/) (Gilson, et al. [35]) databases. These databases were queried using component names or their canonical smiles obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Kim [36]). The screening criteria used for each database were as follows: SEA - Tanimoto coefficient (TC) > 0.5, STP - top 15 protein targets, SP - all known protein targets and > 90% of predicted protein targets, HIT 2.0 - direct & indirect up/down-regulated, TTD - all evidence-based, DCdb - Activity value (-log[M]) > 4.0, DGIdb - interaction score > 0.5, and BindingDB – similarity > 0.85 protein targets. For acquiring allergic rhinitis-related protein targets, two web servers were used: GeneCards (https://www.genecards.org/) (Stelzer G, et al. [37]) and DisGeNET (https://www.disgenet.org/) (Piñero, et al. [38]).
The screening criteria of protein targets for GeneCards were protein-coding genes and a relevance score > 4.0, while for DisGeNET, it was GDA (gene-disease association) score > 0.1. The search keywords used for the allergic rhinitis-related protein target’s acquisition were "seasonal allergy," "hay fever," and "allergic rhinitis." All protein targets were cross-verified using UniProt ID to confirm the correctness of Gene ID, and only human-related protein targets were used for this study. A shared target strategy was employed to establish a relationship between protein targets of AR components and allergic rhinitis. Shared targets were identified using Venny 2.1, available at https://bioinfogp.cnb.csic.es/tools/venny/ (Oliveros Juan Carlos [39]).
Protein-Protein-Interaction (PPI) Network
The shared protein targets were analyzed using the STRING plugin in Cytoscape (Shannon, et al. [40]), a bioinformatics software tool widely used for visualizing network interactions. This facilitated exploring protein target’s interaction via the PPI network using a high confidence score of > 0.7. The interconnected protein targets in the PPI network were considered putative protein targets and subjected to further analysis, while the non-connected protein targets were eliminated.
Acquisition, Enrichment, and Identification of Relevant Functions
In this study, GO (Gene Ontology) (Carbon, et al. [41]) immune system biological processes and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were annotated as ‘Functions.' The modulation of various functions influences the allergic rhinitis condition. The functions related to the screened putative protein targets were extracted using the ClueGO plugin in Cytoscape (Bindea, et al. [42]). The functions were enriched based on a p-value threshold of < 0.05, and only the relevant functions for allergic rhinitis were screened.
Target-Target-Interaction (TTI) Network and Module Generation
A TTI network was constructed to analyze the interconnections among protein targets through various functions. A correlation file for protein targets and their associated functions was generated using Edges 2.0 (E Thorpe, et al. [43,44]). The file was subsequently uploaded to Gephi (Bastian, et al. [45]) to construct an undirected network and identify modules using the Louvain algorithm (resolution 1.0). These modules represent clusters of protein targets linked closely through stronger interrelations (various functions). Calculation of integrated centrality (IC) score and identification of important protein targets. The importance of a protein target was calculated using the integrated centrality equation (X Li, et al. [46]) (Equation 1)
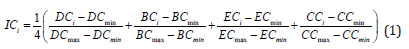
where IC is the integrated centrality score for the protein target (i), and DC, BC, EC, and CC are the degree, betweenness, eigenvector, and closeness centralities, respectively. The max and min represent each centrality's maximum and minimum values within the module. The IC score is a numerical value obtained using the IC equation ranging from 0.00 to 1.00, where 0.00 is the least important and 1.00 is the most important protein target in the module based on the topological perspective.
Molecular Docking Simulations
In-silico validation was performed using molecular docking simulations to elucidate the candidate compound's binding affinity (kcal/mol) towards the protein target. The protein targets with the highest IC score within each module were selected for docking simulation with their respective AR candidate components. The three-dimensional structures of the protein targets and AR candidate components were prepared for docking analysis using PyMOL (https://pymol.org/2/) (Schrödinger LLC, 2015), Open Babel GUI (https://openbabel.org/) (O’Boyle, et al. [47]), and AutoDock 4.2 (https://autodock.scripps.edu/download-autodock4/) (Morris et al. [48]). Once the protein targets and AR components were prepared, they were subjected to docking analysis using AutoDock Vina (Trott, et al. [49]). The protein targets were set as receptors, while the components were ligands. Additionally, docking simulations were performed using reported inhibitory compounds/antagonists of these protein targets to compare and evaluate the efficacy of the AR components based on the binding affinities.
The Workflow
All the components of AR were initially acquired, assessed, and screened for their OB and DL properties. Subsequently, protein targets for the screened AR components and allergic rhinitis were obtained and evaluated. The shared protein targets in AR components and allergic rhinitis were used to construct a PPI network. The PPI network was pruned only to identify the connected protein targets to obtain functions (biological processes and pathways). Functions were enriched based on significance levels (p-value < 0.05) and screened for relevance to immune system diseases, specifically allergic rhinitis. The relevant functions and protein targets were used to construct a TTI network. The TTI network was analyzed to identify pivotal protein targets based on modularity analysis and integrated centrality score using the Louvain algorithm and integrated centrality equation, respectively. The pivotal protein targets and their corresponding AR candidate components were selected using molecular docking simulations to determine the binding affinities. The AR candidate components having the lowest binding affinities towards their respective pivotal protein targets were identified as key bioactive AR components.
Data Acquisition and Screening
Eighty-seven components of AR were acquired through a database search using three web databases (TCMSP, TCM-ID, and ETCM). These components were screened using a threshold of OB > 10% and DL > 0.10, resulting in 40 AR candidate components (Table 1). Using the names and canonical smiles obtained from PubChem for these candidate components, 558 non-repeating protein targets were retrieved from the databases, as mentioned previously. Additionally, a search for allergic rhinitis-related protein targets using different synonyms identified 228 non-repeating protein targets from GeneCards and DisGeNET post- screening. Using a Venn diagram, a shared protein target strategy was implemented to identify protein targets common to both AR candidate components and allergic rhinitis, resulting in 44 protein targets. Table 2 presents these shared protein targets along with their UniProt IDs.
PPI Network Construction
The 44 shared protein targets' PPI network was constructed using the STRING plugin in Cytoscape, forming a network consisting of 249 edges connecting 42 protein targets with an average clustering coefficient of 0.745. However, 2 of the 44 shared protein targets were removed due to the unavailability of their connections with the rest of the network as the set criteria for minimum interactions of a protein target was greater than or equal to 2. The connected 42 putative protein targets were chosen for functional acquisition and enrichment. Figure 1a displays the PPI network of the 42 shared protein targets, with nodes (protein targets) and their edges (interactions via functions).
Enrichment and Identification of Relevant Functions
Functional acquisition of the 42 putative protein targets was performed using Cytoscape, retrieving 140 functions (77 GO biological processes and 63 KEGG pathways). These functions were then enriched based on their significance score (p-value < 0.05) and relevance to allergic rhinitis, resulting in the identification of 42 functions (21 GO biological processes and 21 KEGG pathways). Figure 1b displays the functions and associated genes per term as a percentage. Once the relevant functions were identified, only the protein targets related to the 42 relevant functions were selected for further studies, referred to as candidate protein targets, 32 in this study.
TTI Network Construction and Analysis for Modularity and Important Protein Target Identification
The data set comprising 32 candidate protein targets and 42 functions was used to construct an undirected TTI network. The constructed network consisted of 32 nodes and 319 edges. Four modules were generated using the Louvain algorithm, namely, Module 1 (green color) with 8, Module 2 (purple color) with 9, Module 3 (orange color) with 8, and Module 4 (blue color) with 7 candidate protein targets, as shown in Figure 2. The present study employed IC analysis to identify important protein targets (See Section 2.6). A higher IC score indicates that a protein target has a greater number of connections with other protein targets in the same module via various functions. Candidate protein targets having the maximum IC score (high importance) in each module were selected as pivotal protein targets. ALOX5 (Lipoxygenase-5), IL2 (Interleukin-2), TNFα (Tumor necrosis factor-α), and TLR4 (Toll-like receptor 4) had the highest IC score in modules 1, 2, 3, and 4, respectively. The modules and their corresponding IC scores are listed in Table 3.
Molecular Docking Simulations
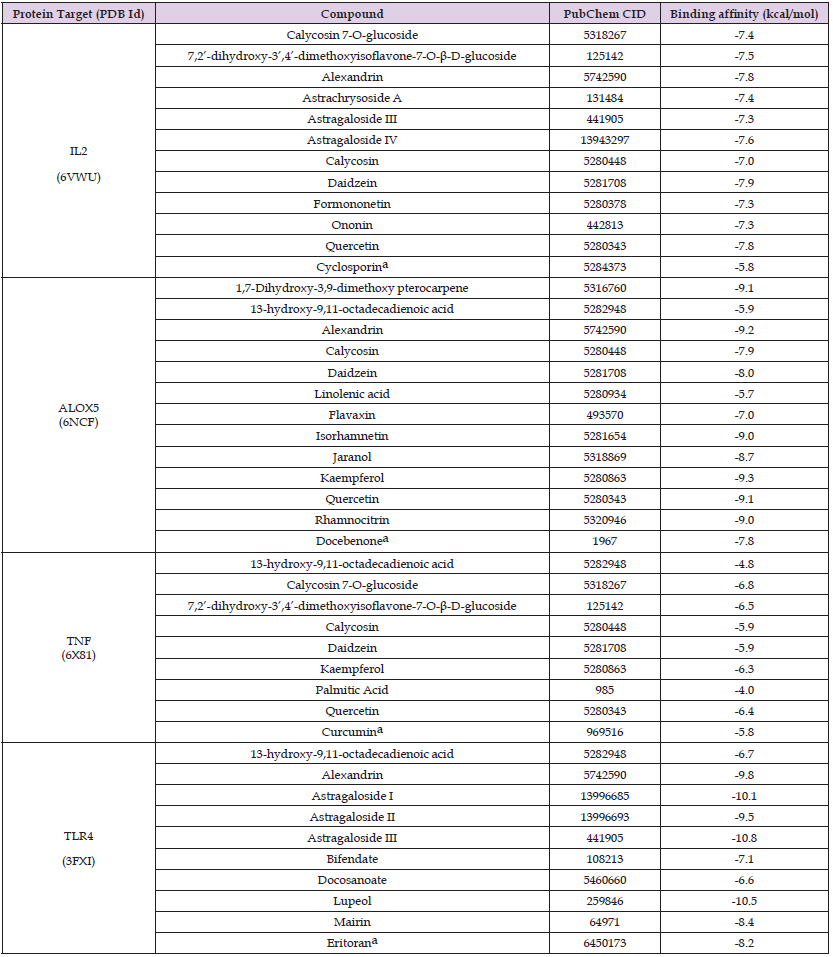
Molecular docking simulations were performed on the pivotal protein targets and their related AR candidate components, as shown in Figure 3. The binding affinities obtained for each protein-ligand complex are mentioned in Table 4. Based on the docking results, the AR candidate components showing the best binding affinities towards ALOX5, IL2, TNFα, and TLR4 were kaempferol (-9.3 kcal/mol), daidzein (-7.9 kcal/mol), calycosin 7-O-glucoside (-6.8 kcal/mol), and astragaloside III (-10.8 kcal/mol), respectively and were selected as the key bioactive AR components. It exhibits that these key bioactive AR components can be used to regulate the pivotal protein targets, as they showed excellent binding scores, with values less than -5.0 kcal/mol (B Li, et al. [50,51]); Additionally, the binding affinities of these key bioactive AR components were compared with the reported inhibitory compounds/antagonists for each protein target (Figure 4). It was observed that the key bioactive AR components had much lower (better) binding affinities than the reported inhibitory compounds/antagonists for TNFα, IL2, ALOX5, and TLR4. Furthermore, the binding pockets of the key bioactive AR components were the same as those reported for inhibitory compounds/antagonists, predicting the antagonist action of the key bioactive AR components.
Table 4: Binding affinities of the vital protein targets to the candidate components of A. radix and their reported inhibitory compounds/antagonists.
This study embarked on a quest to delineate the bioactive components of AR and their significant protein targets, offering therapeutic potential against allergic rhinitis. Utilizing network pharmacology, we intricately mapped the interplay between these components and protein targets, employing the scientific findings of chemometrics and bioinformatics. A pivotal aspect of our methodology was constructing the TTI network, which revealed the intricate web of connections between proteins across various biological processes and pathways. A core challenge in network pharmacology is pinpointing the most influential protein targets within a vast network. Our study addresses this through modularity analysis and integrated centrality scoring. We identified modules – the subnetworks of interlinked protein targets - and ranked the protein targets in each module using integrated centrality scores. This dual strategy facilitated the discernment of protein targets, wielding an understanding of the molecular action mechanism of the bioactive components of AR, a task often convoluted in complex biological networks. Molecular docking emerged as a crucial tool, enabling us to scrutinize the interactions between AR components and protein targets. This approach was instrumental in revealing potential inhibitory or activating binding sites and energies, thereby guiding therapeutics’ development. Our research spotlighted four pivotal protein targets: ALOX5, IL2, TNFα, and TLR4. These proteins play a distinctive role in immune response and allergic pathophysiology.
For instance, when activated inappropriately, TNFα, is integral to immune system balance and can exacerbate allergic rhinitis via the NFκB pathway (Ahmad, et al. [6,52]). Similarly, TLR4's role in initiating inflammatory responses (Radman, et al. [53,54]) and ALOX5's involvement in leukotriene biosynthesis (Çobanoğlu, et al. 55,56) underscore their relevance in allergic reactions. This study uniquely considered the collective impact of these targets, a perspective often overlooked in existing literature. Beyond confirming known pathways implicated in allergic rhinitis, our research ventured into previously unexplored territories. We unearthed additional pathways and biological processes influenced by AR, such as the PI3K-AKT and MAPK signaling pathways, thereby broadening our understanding of allergic rhinitis pathogenesis. In-silico validation pinpointed four AR components - calycosin-7-O-glucoside, kaempferol, daidzein, and astragaloside III - as potential therapeutic agents. These compounds, notably flavonoids and triterpenoid saponins, exhibited promising binding scores and specific biological activities against allergic inflammation (Jia, et al. [57-62]). This raises an intriguing question about their collective versus individual efficacy in modulating allergic responses. The findings of this study underscore the therapeutic promise of AR's key bioactive components in modulating pivotal protein targets and pathways in allergic rhinitis. However, these preliminary insights beckon further investigation to fully elucidate their clinical potential and the intricacies of AR's multi-component synergy. Future studies should focus on validating these results and exploring their practical applications in allergy treatment.
This research leveraged the power of network pharmacology to unravel the molecular mechanism of Astragalus radix for the treatment of allergic rhinitis. At its core, this study identified a suite of bioactive components in AR and their specific disease protein targets, shedding light on their therapeutic potential. These findings were bolstered by in-silico validation through molecular docking simulations, corroborated by scientific literature. This research marked a noteworthy leap in connecting traditional herbal medicine knowledge with the understanding of disease genes, filling a crucial gap in mechanism-focused studies. Our method simplifies the traditionally demanding process of pinpointing and testing protein targets and active ingredients. This study provides verifiable mechanisms and paves the way for more effective clinical trials targeting AR’s active components for allergic rhinitis treatment.
The authors thank the Department of Chemistry at Cleveland State University.
The authors declare that they have no conflict of interest.
The datasets used during the current study are available from the corresponding author upon reasonable request.
ASP performed data acquisition, network pharmacology, data analysis, and molecular docking and drafted the manuscript. YX designed and supervised the study and made necessary revisions to finalize the manuscript. All authors have read and agreed to the published version of the manuscript.


