Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Kewei Zhang1,2* and Thushari Indika Alahakoon2,3
Received: June 09, 2023; Published: July 06, 2023
*Corresponding author: Kewei Zhang, Westmead Institute for Maternal Fetal Medicine, Western Sydney Local Health District Research, Education and Network Building, Department of Obstetrics and Gynaecology, Westmead Hospital, Corner of Hawkesbury and Darcy Roads, Westmead, New South Wales, 2145, Australia
DOI: 10.26717/BJSTR.2023.51.008100
Background: Autosomal dominant tubulointerstitial kidney disease (ADTKD) is a rare cause of end-stage renal failure (ESRF), mutations of which primarily affect UMOD (ADTKD-UMOD) gene. Over 120 mutations have been identified and the majority are clustered in UMOD exons 3 and 4. Some mutations have been found with high frequency in families outside Australasia; however, diagnosis of mutations in Australian families with ADTKD is uncommon. The aim of this study was to examine mutations in UMOD in five Australian families with ADTKD and compare with literatures.
Methods: We examined all members of five Australian families with ADTKD for UMOD mutations by amplifying exons 3 and 4 and direct sequencing. Genomic DNA was isolated from peripheral blood or buccal swabs from kindreds.
Results: Complex heterozygous in-frame indel mutation, c.278_289delTCTGCCCCGAAG/ insCCGCCTCCT, was found in exon 3 in a multigenerational Australian family. This mutation resulted in the replacement of five amino acids (Val93CysProGluGly97del) by four new ones (insAlaAlaSerCys) and segregated with the disease. A second heterozygous substitution, c.854C>A, was identified in UMOD exon 3 in another Australian family. This mutation resulted in an Ala285Glu change that segregated with disease. The two mutations were not present in 50 normal controls.
Conclusion: This study first identified two mutations clustered in UMOD exon 3 in two Australian families. These two recurrent UMOD mutations, previously reported with high frequency in patients with ADTKD in European and American families, were possibly due to a founder effect. Routine UMOD mutational testing should be considered for patients with ADTKD in Australia.
Keywords: Uromodulin; UMOD; ADTKD; Founder Effect
Abbreviations: ADTKD: Autosomal Dominant Tubulointerstitial Kidney Disease; ESRF: End-Stage Renal Failure; ESRD: End-Stage Renal Disease; MUC1: Mucin-1; REN: Renin; HNF1ß: Hepatocyte Nuclear Factor-1 ß; MCKD2: Medullary Cystic Kidney Disease Type 2; FJHN1: Familial Juvenile Hyperuricemic Nephropathy Type 1; UAKD: UMOD- Associated Kidney Disease; EGF: Epidermal Growth Factor; cbEGF: Ca2+ Binding Consensus of EGF; GPI: Glycosyl Phosphatidy Linositol; CKD: Chronic Kidney Diseases; PCR: Polymerase Chain Reaction; ER: Endoplasmic Reticulum
ADTKD, a rare cause of end-stage renal disease (ESRD), is characterized by tubular damage and interstitial fibrosis primarily due to mutations in UMOD, and mucin-1 (MUC1) genes. Uncommon forms of ADTKD have also been associated with mutations in renin (REN), hepatocyte nuclear factor-1 ß (HNF1ß) and SEC61A1 genes [1,2]. ADTKD-UMOD, the most common form, was previously known as the allelic disorders: medullary cystic kidney disease type 2 (MCKD2), familial juvenile hyperuricemic nephropathy type 1 (FJHN1), or as UMOD-associated kidney disease (UAKD) [1,3,4]. MUC1 and REN mutations cause fewer forms of MCKD1 and FJHN2 separately [1]. The clinical features of ADTKD are hyperuricemia, gout, and hypertension with variable penetrance, which eventually develop into ESRD. The absence of hematuria and proteinuria, however, is noteworthy. The nonspecific clinical presentations are often delayed, and renal biopsy features such as tubular atrophy and medullary cysts are not specific for all patients with ADTKD-UMOD [1,5]. The UMOD gene, first mapped to 16p12.3, is made up of 11 exons and encodes the uromodulin (Tamm-Horsfall), which is the most abundant protein in urine [6,7]. Uromodulin consists of 640 amino acids containing an N-terminal signal peptide (mainly exon 3), and three epidermal growth factor (EGF) domains, with Ca2+ binding consensus in two of EGF (cbEGF2 and cbEGF3). This is followed by a sequence including domain of 8 cysteine residue that is highly conserved in different species. A fourth cbEGF-like domain extends through exon 4 and is then followed by a zona pellucida at the C-terminal region and glycosylphosphatidylinositol (GPI) anchor site [2,5,8,9]. Families with ADTKD-UMOD mutations have been reported in Europe, the USA, Asia, and Africa [10-12] but not in Australia. Over 120 mutations have been described in ADTKD-UMOD and above 93% clustered in exons 3 and 4 where they affect the cbEGF-like domains [1,2,5,13,14]. Most UMOD mutations are missense mutations, resulting in the replacement of cysteine residues [2]. This leads to Endoplasmic Reticulum (ER) retention of mutant uromodulin within tubular cells, misfolding and excreting less wild-type uromodulin, thus causing tubulointerstitial damage [4,9,14]. A genome-wide association study has identified some variants in UMOD gene promoter as risk factors for chronic kidney diseases (CKD), suggesting that the level of uromodulin in the urine could be a marker for CKD [9,15-17]. Recent study shows that 3% of patients with a monogenic cause of CKD had UMOD mutations [2]. Clinicians under-recognize familial clustering among patients with CKD, of which potential ADTKD-UMOD might be the most frequent [18]. Recently, genetic testing has emerged as a diagnostic option for ADTKD [1,8]. Australian clinicians underutilize genetic testing for ADTKD, hence mutational detection is not routinely performed. The aims of this study were to examine mutations in exons 3 and 4 of UMOD gene in Australian families previously diagnosed with ADTKD, and to compare UMOD mutations with the literature. To our knowledge, we describe the first search for mutations in UMOD gene in Australian families who have ADTKD and ESRF without pronounced hematuria or proteinuria.
Patients’ Information
We studied five unrelated index cases and other members from five Australian families (Table 1). All five index cases had renal impairment but minimal hematuria and proteinuria, and the clinical features were consistent with the diagnosis of ADTKD. All five had renal biopsy confirmation of tubulointerstitial disease. Five indexes were Caucasian; four were European, of which kindred one was Welsh, and one was Italian. We collected blood and urine for all five indexes and other members from five families.
Note: *Central serous retinopathy; fundus albipunctatus; optic nerve meningioma; retinal scarring (4 patients examined). M: male; F: female; AD: autosome dominant; U/K: unknown. N/A: not available.
Mutation Detection in UMOD
We extracted genomic DNA from peripheral blood or buccal swabs as described previously [19]. Only exons 3 and 4 of UMOD were amplified and examined for mutations using specific primers that were designed to divide these exons into three overlapping fragments (Table 2 & Figure 1). We designed three pairs of primers, which divided exons 3 and 4 into three overlapping parts. The first part of exon 3 included 3.1 forward (F) and 3.1 reverse (R) (504bp); the second part of exon 3 was 3.2F and 3.2R (517bp); and the third part of overlapping exons 3 and 4 was 3.4F and 4R (320bp). The first part of exon 3 was amplified twice by nested polymerase chain reaction (PCR). The first-round PCR of the first part in exon 3 was amplified using 3.1F and 3.2R primers (965bp). The second round was subsequently amplified using 3.1F and 3.1R primers (504bp) and first- round product as template. The second part of exon 3 was also amplified using nested PCR. The first-round PCR of the second part in exon 3 was amplified as above, using 3.1F and 3.2R primers (965bp). The second round was subsequently amplified using 3.2F and 3.2R primers (517bp) and first-round product as template. The last part was amplified to overlap exon 3 and 4 using 3.4F and 4R primers (320bp) (Figure 1). The amplification conditions were shown in Table 3. The amplification products were purified and sequenced in both directions using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PerkinElmer) and an ABI automated DNA sequencing machine. We examined sequence variants to determine whether they had been reported previously as mutations and whether they changed an amino acid, particularly conserved residue across at least three different organisms (mouse, dog and cow mRNA UMOD sequences); whether they segregated with disease where DNA from family members was available; and whether they were present in 50 normal controls.
Note: F: forward; R: reverse.
Figure 1 Primers sequence in exons 3 and 4 in the UMOD gene Three pairs of primers (3.1F-3.1R, 3.2F-3.2R, and 3.4F-4R) were designed to amplify 3 and 4, and one pair of primers (3.3F-3.1R) was designed to detect the complex mutation (c.297del12/ins9).

Note: PCR: polymerase chain reaction; bp: base pair.
Family Analysis
Thirty members of kindred 1 and nine members of kindred 2 were studied to determine whether the presence of UMOD mutations segregated with disease (the pedigree of kindred 1 was not available). To confirm the complex mutation, a new forward primer (Exon3- 3.3F) and reverse primer (Exon3-3.1R) (Figure 2) were designed to amplify short PCR product (230bp) in the first part of the exon 3. The products then underwent electrophoresis in a 2% agarose gel. The presence of this mutation with double band was then correlated with the clinical features.
Figure 2 A. A c.278_289del12/ins9 mutation in UMOD gene in the Family 1 with ADTKD in reverse sequencing A heterozygous 12-nucleotide deletion and an insertion of nine nucleotides (arrow) in exon 3 of the UMOD gene in reverse sequence in Family 1 resulted in the replacement of five amino acids (Val93CysProGluGly97del) by fourdifferent ones (insAlaAlaSerCys). B. 2% Agarose gel electrophoresis of exon 3 in UMOD gene in seven index cases (1 to 8) of Family 1 Double bands at 230 bp showed there was a heterozygous mutation (c.279del12/ins9) in each affected individual, pUC19: DNA/MspI molecular marker, sample 1 and 2 are duplicates.

Clinical Data
All five patients had renal impairment but minimal hematuria and proteinuria, and other clinical features were consistent with the diagnosis of ADTKD. All three patients for whom urate levels were measured had hyperuricemia. The median age at onset of kidney failure was 32 years (range, 20–50 years). We also performed ocular examinations for four patients, as described in a previous study [19]. Three (3/5, 60%) had normal ocular examination and one patient in Family 1 (1/5, 20%) had various inconsistent ocular abnormalities such as central serous retinopathy, fundus albipunctatus, optic nerve meningioma, and retinal scarring. All five patients (5/5,100%) had renal biopsy confirmation of tubulointerstitial disease (Table 1). Four individuals (4/5, 80%) had at least one other known affected family member. Inheritance in these families was autosomal dominant with multiple affected male and female family members. Similar sex and severity distributions were noted in the four families. One patient (from Family 5) did not have another affected family member. All other members of two multigenerational families 1 and 2 with mutations were further studied for whether mutations segregated with disease.
Mutation Detection in UMOD
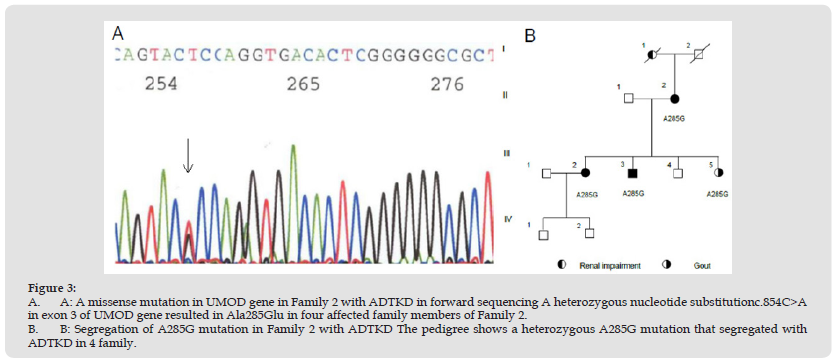
We sequenced DNA samples from five indexes and other family members. Two mutations were identified in affected indexes from Families 1 and 2. The mutation in Family 1 was an in-frame, heterozygous 12-nucleotide deletion with an insertion of nine nucleotides of c.278_289delTCTGCCCCGAAG/insCCGCCTCCT (c.278- 289del12/ ins9) in forward sequence in exon 3, resulting in the replacement of five amino acids (Val93CysProGluGly97del) by four different ones (insAlaAlaSerCys) in forward sequence ((Figure 2a) shows the complex mutation in reverse sequence). The heterozygous c.278_289del12/ ins9 mutation was evident as a double band on 2% agarose gel electrophoresis (Figure 2b). It was present in the seven affected members of Family 1 who were tested but was not present in the 23 unaffected members, and thus segregated with disease (Family 1 pedigree was not shown). This mutation was not found in any of 50 normal individuals without hematuria or proteinuria. Another mutation in Family 2 was a missense heterozygous nucleotide substitution c.854C>A in exon 3 that resulted in an Ala285Glu change (Figure 3a). This mutation also affected the EGF-like domain of UMOD. It segregated with disease within Family 2 (Figure 3b) and was not found in 50 normal individuals. No novel polymorphisms were identified in any of the five individuals studied.
Figure 3 A. A: A missense mutation in UMOD gene in Family 2 with ADTKD in forward sequencing A heterozygous nucleotide substitutionc.854C>A in exon 3 of UMOD gene resulted in Ala285Glu in four affected family members of Family 2. B. B: Segregation of A285G mutation in Family 2 with ADTKD The pedigree shows a heterozygous A285G mutation that segregated with ADTKD in 4 family.
ADTKD is a rare kidney disease underdiagnosed due to nonspecific clinical features and renal biopsy. Genetic mutational detections for ADTKD are not routinely performed in Australia. We identified two mutations in exon 3 of UMOD in two different Australian multigenerational families with ADKTD. These two mutations in Australian Families 1 and 2 affected amino acids in the highly conserved EGF-like domains of UMOD, which confirm the specific role of exon 3-encoded sequence in the generation of the ADTKD renal phenotype [13]. The complex heterozygous c.278_289del12/in9 mutation affected the second cbEGF-like domain of the uromodulin protein. The functional characterization of c.278_289del12/in9 revealed retarded intracellular trafficking associated with endoplasmic reticulum (ER) retention and reduced secretion into cell culture media [8]. Two of the five amino acids involved in this mutation were highly conserved in evolution across species, including the lower vertebrate zebrafish and the nematode Caenorhabditis elegans, in genes encoding EGF-binding domains [13]. c.278_289del12/ins9 mutation has been described in several human diseases, including Marfan syndrome, where they involve the same mcbEGF domain in FBN1 as in the Family 1 mutation described here [20-23]. The c.278_289del12/ins9 mutation in Family 1 involved a cysteine residue. Cysteines are usually highly conserved in the uromodulin protein. Uromodulin comprises 48 cysteines per monomer that form 24 intramolecular disulfide bonds. The amino acid replacement in c.278_289del12/ins9 changes the molecular conformation through impaired intra- or intermolecular disulfide bond formation [13]. The high cysteine content of uromodulin and the correct formation of the disulfide bonds are thought to be the rate-limiting factors for the export of the premature protein out of the ER. This is thought to regulate the efficiency of uromodulin protein maturation [23]. Until now, most UMOD mutations have been identified in exons 3 and 4, and most mutations are in front of the zona pellucida structure [13]. We review all cases with indel from literature and make a summary. The complex c. 278_289del12/ins9 heterozygous mutation in Australian Family 1 (originating from Welsh), described previously in one large four-generation Welsh family, other four different British families and one European/American family, as well as in a cluster of British families by 4 different groups [2,8,18,24], may be due to a founder event. Interestingly, in four different British families with mild ADTKD, c. 278_289del12/ins9 showed homozygous status. No shared common haplotype was found for this complex mutation. There was no information regarding which microsatellite markers had been used for haplotype analysis [8]. However, Valluru and colleagues recently reported the same UMOD c. 278_289delTCTGCCCCGAAG insCCGCCTCCT mutation was detected in a cluster of unrelated British families which widely distributed within the UK population and all cases were of white British ancestry. Also, the patients with this complex mutation shared expended haplotype. These observations were consistent with a founder effect [24]. But we suggested the founder effect hypothesis should be also further investigated by a population genetic test [25] for this complex mutation with high frequency in British population. Fewer than 2000 families with ADTKD- UMOD have been identified worldwide [26]. We hypothesize that unusual high frequency allele in British-Welsh, European-American and Australian-Welsh families could result from the establishment of a new population from individuals who carried the same mutation derived from a larger population (the British-Welsh populations). Alternatively, the allele could result from an extreme reduction in population size, such as the population ‘bottleneck’ effect. In either case, the allele present in one copy after the founder or bottleneck event may be found at a higher frequency than it was previously and could reach higher frequencies because of genetic drift occurring while the population is small [27]. Further linkage analysis using the same microsatellite markers could be done for all affected British-Wales, European/American and Australian families to determine whether this complex mutation is due to a founder event or just because the particularly local DNA-sequencing environment (exon 3) creates a mutational hotspot The missense heterozygous nucleotide substitution c.854C>A in exon 3 that resulted in an amino acid change Ala285Glu in Family 2 has also been reported previously but no patient information and family place of origin were described in detail [14]. Bollee’s group identified the c.854C>A (Ala285Glu) mutation in French and Belgian families [28]. We hypothesized that this mutation might be also due to a founder effect of European origin. To prove this, further studies will need to be conducted using haplotype analysis for the two different European families and Australian family 2 with c.854C>A mutation. We did not find mutations in UMOD exons 3 and 4 in another three individuals with ADTKD in our study. They may have mutations located in other exons of the UMOD or in other genes e.g., MUC1, REN, HNF-1β or SEC61A1, because the phenotypes of these diseases are clinically similar. Patients with ADTKD usually have milder disease, with less hyperuricemia and gout, and with later onset of ESRF. Approximately 40-75% of analyzed families have disease due to a gene other than UMOD [12,13, 29]. The MUC1 mutations are another most probable cause of ADTKD and occurred in 64% of the families in one study [30]. A further gene locus has been identified at 1q41 in a single Belgian family with features like ADTKD, but for whom the gene REN has been excluded [31]. Possible mutations have been described in three different genes at this locus (AK000210, CCT3 and SCAMP3) but no single causative gene has been identified. A fourth locus is possible [31]. One of the patients from Family 5 that we studied here may not have had ADTKD, because his clinical features were atypical and there was no family history of renal failure. In addition to the two mutations (c.854C>A and c.278_289del12/ins9) identified in the two Australian and European families, the literature also demonstrates another founder effect which is heterozygous c.774 C/G (Cys248Trp) and A285E mutations, by different research groups [28]. This mutation has been identified in six family members from two unrelated families in European Turkish, and Chinese families, and haplotype analysis showed co-segregation with the phenotype in all affected families [32-35]. In Wolf’s study, the microsatellite D16S3054, with different alleles presented in different kindreds, could be attributed to an ancestral mutation of UMOD in the repeat of the microsatellite marker [32]. ADTKD gene mutations are a cause of kidney failure in the Australian community and may have been carried by early European settlers in Australia. The disease has continued because affected individuals were not diagnosed until after they had another affected member in the families. There are no reports regarding mutational detection for families with ADTKD in Australia. Clinicians may under-recognize this condition. No studies investigate UMOD or other genetic mutations as causes of ADTKD in Australia. Our review on all the mutations identified in UMOD in literature shows for the first time that ADTKD in Australia can be due to two recurrent genetic mutations. We suggest that routine mutational detection for familial ESRF or CKD would be useful to find the causes of the condition at an early stage. The critical role of genetic testing for ADTKD is that it allows family members who qualify as donors and wish to donate kidneys to be found. Furthermore, as ADTKD affects generations of families mutational detection may be used to provide both genetic counselling for these families, and potential treatments of the disease.
We gratefully acknowledge the contributions of Professor Judy Savige for introducing the patients to the study, and Associate Professor Deborah Colville (the University of Melbourne) for performing the ocular examinations. We were also grateful that Olivia Wroth (Western Sydney Local Health District Research and Education Network Building) reviewing this paper.
This study was supported by the National Health and Medical Research Council of Australia, Kidney Health of Australia, and Australian Postgraduate Award Scholarship, the University of Melbourne between 2003 and 2007.
The authors have no interests to disclose.
This study design, material preparation, experimental performance, data collection and analysis were performed by Kewei Zhang. The first draft of the manuscript was written by Kewei Zhang. Thushari Indika Alahakoon provided advice on the validity of the content of this paper and reviewed as well as corrected the draft. All authors read and approved the final manuscript.
This study was approved by the Human Research Ethics Committee of Austin and Northern Health, the University of Melbourne and with the principles of the Declaration of Helsinki 1964 and its later amendments or comparable ethical standards.
Informed consent was obtained from all individual participants included in the study.


