Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Alexander J Burdette*, Hector Lopez, Brandie K Goren and Susana N Asin
Received: June 12, 2023; Published: June 26, 2023
*Corresponding author: Alexander J Burdette, Office of Science & Technology Center for Advanced Molecular Detection, Air Force 59th Medical Wing, 2460 Pepperell Street, Building 4429, JBSA Lackland, USA.
DOI: 10.26717/BJSTR.2023.51.008058
The SARS-COV-2 pandemic has had severe world-wide consequences on human and economic health. Rapid diagnosis is critical to prevent transmission. This study evaluates two fully integrated sample processing platforms for SARS-COV-2 detection in saliva. The Xpert® Xpress CoV-2/Flu/RSV plus cartridge is used on the laboratory-based Cepheid® platform, while the XCEL™ Respiratory ISP cartridge is used on the portable Franklin® ISP system. SARS-COV-2 patient research saliva samples (150 negative; 124 positive) were collected the same day as the nasal swab sample for the official CLIA test allowing for direct comparison. The limit of detection in saliva for the Cepheid® cartridge was 8400 copies/mL, while the Franklin® ISP cartridge was over 42,000 copies/mL. The Cepheid® cartridge exhibited sensitivity/specificity of 56.56%/98.11%, while the Franklin® ISP system was 28.72%/94.56%. While both systems had high specificity for SARS-COV-2 detection in saliva, the sensitivity was low, thus requiring further refinement of the systems for saliva detection.
Keywords: SARS-COV-2; Saliva; Biomeme; Cepheid®
Severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) began in China in late 2019. Since then, it has spread to other countries creating a world-wide pandemic with multiple variants. Symptoms can range from asymptomatic, to severe symptoms with hospitalization including ventilation and death [1-3]. Diagnostic kits have quickly arisen to address the SARS-COV-2 pandemic. These kits include systems such as real-time polymerase chain reaction (RT-PCR), isothermal based PCR and lateral flow strips for IgG/IgM antibody detection [4-6]. While nasopharyngeal swabs have predominantly been the gold standard for sample collection and testing using RT-PCR for SARS-COV-2, the collection procedure requires specialized training and can be painful for the patient [7,8]. Further, due to the specialized training to collect the nasopharyngeal swab samples, possible SARS-COV-2 infected patients have to come to patient waiting rooms in emergency clinics and hospitals to give their samples, which can lead to further infection transmission to hospital providers and other patients.
Anterior nasal swabs do not require specialized training to collect, but the patients do have to ensure they are adequately swabbing their nose to ensure the virus, if present, is collected to avoid a false negative test. Saliva however, is a simple specimen source that requires no training to collect nor concern for adequate swabbing by the patient, and is non-invasive. In the civilian setting, because no training is required to collect saliva, this sample source could be collected at home and then dropped off at a drive-thru for actual testing. Importantly, studies have validated that SARS-COV-2 is detectable in saliva [9-14].
One portable RT-PCR system that could be of use in different environments where space is limited such as drive-thru testing facilities, are the Franklin® RT-PCR systems (Biomeme, Philadelphia, PA). These RT-PCR systems are small, ruggedized, lightweight systems that pro vide results via Bluetooth through a smartphone and can run up to 27 targets per run. Our lab has previously worked with the Franklin® RTPCR system which utilizes RT-PCR test strips. However, the device and RT-PCR kits require a separate step and separate device for RNA isolation. The system was found though to be effective for SARS-COV-2 detection [15]. However, a more recent system has been developed which is a fully integrated sample processing (ISP) and detection system (Franklin®ISP) with an internal battery wherein RNA isolation, conversion to cDNA amplification and detection are all automated and take place within the same cartridge. In combination with saliva as a sample source, this ISP system could provide an ideal SARS-COV-2 detection system for use in drive-thru testing facilities, and other environments where space and power is limited. In this study, we evaluate and compare the sensitivity and specificity of SARS-COV-2 in saliva of the Franklin® ISP systems with the XCEL™ Respiratory ISP cartridge, to the FDA approved Cepheid® ISP systems with Xpert® Xpress CoV-2/ Flu/RSV plus cartridge.
Clinical Samples
This study was approved under the 59th Medical Wing Institutional Review Board protocol FWH2020087N. Saliva samples (n=150 SARS-COV-2 negative; n=124 SARS-COV-2 positive) were commercially purchased from the company iSpecimen (Lexington, MA). Samples were collected on the same day as the clinical nasal swab collected by hospital staff for the official SARS-COV-2 diagnosis at the CLIA lab using the QIAstat-Dx Respiratory SARS-CoV-2 Panel (Redwood City, CA) which served as the Gold Standard. Saliva samples were collected using the Saletto™ Oral Fluid Collection device (Porex Life Science Institute, Fairburn GA) according to manufacturer instructions in a volume of 1mL per patient. Saliva samples from each patient were frozen down in liquid nitrogen in cryotubes and shipped to our research facility. Once received, samples were aliquoted into Eppendorf tubes for two different PCR systems being evaluated: the FDA approved Xpert® Xpress CoV-2/Flu/RSV plus on the Cepheid® GeneXpert® Xpress system (Cepheid®, Sunnyvale, CA), and the XCEL™ Respiratory ISP Cartridges on the Franklin® ISP system (Biomeme, Philadelphia, PA). Patient Demographics are described in Table 1.
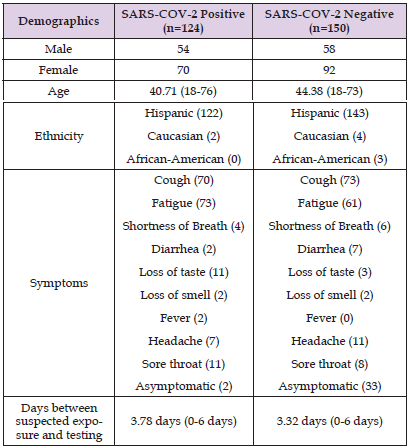
Table 1: Patient Demographics. Shown in the table are the patient factors including gender, ethnicity, symptoms present at time of testing, and days between the suspected exposure and actual testing. Many of the patients had more than one symptom at a time.
Limit of Detection Studies
Limit of detection (LoD) studies were performed with saliva samples for both systems. Serial dilutions of Heat Inactivated 2019 Novel Coronavirus (ATCC® VR-1986HK™; Manassas, VA) were prepared in RNase-free water from a stock concentration of 4.2 × 108 genome copies/ mL. Briefly, 50 SARS-COV-2 negative saliva samples were pooled and then diluted 1:1 into DNA/RNA Preservation Buffer (Biomeme, Philadelphia, PA) for the XCEL™ Respiratory ISP Cartridges. Samples tested on the Xpert® Xpress CoV-2/Flu/RSV plus cartridges did not require a dilution. The concentrations tested for the LoD studies included 42,000 copies/mL, 8,400 copies/mL, 1,680 copies/mL, 336 copies/mL, 67.2 copies/mL at n=6 replicates per concentration. Samples were run on both systems according to manufacturer instructions using 300ul of sample for the Xpert® Xpress CoV-2/Flu/RSV plus cartridges, and 400ul of sample/preservation buffer for the XCEL™ Respiratory ISP Cartridges.
System Comparison Studies
To compare the two systems for sensitivity/specificity for saliva, 150 SARS-COV-2 negative saliva samples and 124 SARS-COV-2 positive saliva samples (based on the result of the official CLIA test) were evaluated on both systems according to manufacturer instructions using 300ul of sample for the Xpert® Xpress CoV-2/Flu/RSV plus cartridges, and 200ul of sample for the XCEL™ Respiratory ISP Cartridges plus 200ul of DNA/RNA preservation buffer.
Statistical Analysis
For the LoD studies, the LoD was determined to be the lowest concentration where 100% of all samples tested positive. Statistical analyses were performed using Graphpad prism software (Boston, MA). Sensitivity and specificity for each system was determined based on the results of the Gold Standard (CLIA test result). For comparative analyses between systems, we used Cohen’s kappa statistics to estimate agreement and test the null hypothesis that agreement was random (i.e., kappa statistic equals zero). Values that are ≤ 0 suggest no agreement, 0.01-0.20 as none to slight agreement, 0.21–0.40 as fair agreement, 0.41– 0.60 as moderate agreement, 0.61-0.80 as substantial agreement, and 0.81–1.00 as almost perfect agreement [16]. It should be noted that these interpretations of agreement are subjective, but for the purpose of this study, these are the interpretations being utilized. McNemar’s chi‐square test was used to test the null hypothesis that the systems are equivalent in regards to sensitivity and specificity (p<0.05). To correlate sensitivity of each system to number of days between suspected onset of SARS-COV-2 and the CLIA test sample/research saliva sample collection, the percent positive samples was determined for each system for CLIA positive samples only for number of days from suspected onset of disease to sample collection date to include 0 days, 1 day, 2 day, 3 days, 4 days, 5 days, 6 days.
Patient Demographics
In this study, we compared the performance of two fully integrated sample processing RT-PCR systems using saliva as the sample source. Table 1 displays the results of the patient demographics. The number of females in both groups were slightly higher than the number of males. Hispanic ethnicity was the overwhelming majority for both groups. The average age for both groups was similar with 40 years old being the average age in the SARS-COV-2 positive group, and 44 years old being the average age in the SARS-COV-2 negative group. Symptoms were varied in both groups. The only symptom that was present in the SARS-COV-2 positive group that was not present in the SARS-COV-2 negative group was a fever (n=2 patients). Cough and fatigue made up the majority of symptoms with most patients presenting with at least those two symptoms. Interestingly, neurosensory symptoms such as loss of taste and smell, were present in both groups.
Limit of Detection
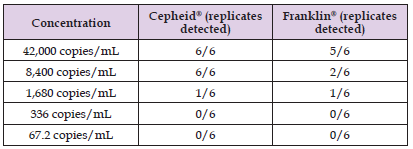
The Limit of Detection (LoD) in saliva was next assessed in both the systems by spiking inactivated whole SARS-COV-2 pathogen into negative saliva samples at five-fold dilutions from 42,000 genomic copies/mL to 67.2 genomic copies/mL. The Cepheid® GeneXpert® Xpress system demonstrated the greatest sensitivity in saliva compared to the Franklin® ISP system with an LoD of 8,400 genomic copies/ mL (100% detection of all replicates). At concentrations lower than 8,400 genomic copies/mL, it quickly lost sensitivity, with only about 16% detection at 1,680 genomic copies/mL and no detection at concentrations lower than that (Table 2). The LoD of the Franklin® ISP system in saliva was higher than 42,000 genomic copies/mL, as only 84% detection occurred at 42,000 genomic copies/mL. The sensitivity of the system quickly dropped with 33% detection at 8,400 genomic copies/mL and 16 % detection at 1,680 genomic copies/mL (Table 2).
Table 2: Limit of Detection (LoD) Experiments. Inactivated SARSCOV- 2 whole pathogen was diluted in saline to the genomic copies/ mL shown in the Table. Six replicates were done per concentration. Shown in the Table is the number of replicates for each concentration that tested positive. The LoD was the lowest concentration where all six replicates were positive.
Performance With Respect to Sensitivity/Specificity and Agreement in Saliva
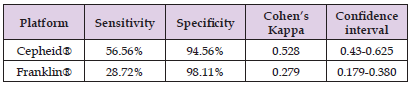
The sensitivity and specificity of each system in saliva was compared. For this experiment, the patient saliva samples were collected at the same time as the nasal swab that was collected for the official SARS-COV-2 CLIA Diagnostic Test. This allowed for a direct comparison to the Gold Standard to determine sensitivity and specificity in saliva of these two systems. Table 3 summarizes sensitivity, specificity and Cohen’s Kappa between each of the systems and the Gold Standard. The Cepheid® GeneXpert® Xpress system demonstrated higher sensitivity than the Franklin® ISP system (56.56% vs 28.72% for both systems), while the Franklin® ISP system performed slightly better than the Cepheid® GeneXpert® Xpress system for specificity (98.11% vs 94.56% for both systems). The overall agreement of the two systems with the SARS-COV-2 CLIA test result was determined by Cohen’s Kappa. As shown in Table 3, The Cepheid® GeneXpert® Xpress system exhibited moderate agreement with the result of the SARSCOV- 2 CLIA Test result at a Cohen’s Kappa of 0.528 (95% Confidence interval 0.43-0.625). The Franklin® ISP system did not have as high an agreement demonstrating fair agreement with the SARS-COV-2 CLIA Test result with a Cohen’s Kappa of 0.279 (95% Confidence Interval 0.179-0.380).
Table 3: Sensitivity and Specificity of Cepheid® and Biomeme Franklin ® Systems Compared to Gold Standard. The Gold Standard in this study is the nasal swab that was taken for the official CLIA Diagnostic Test by hospital staff. The research saliva samples were taken the exact same day to allow for direct comparison to the Gold Standard. Shown in the Table is the sensitivity, specificity and Cohen’s Kappa of the Cepheid® and Biomeme Franklin® systems compared to the Gold Standard.
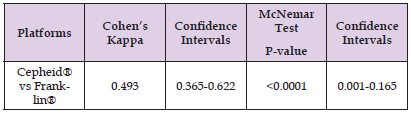
Next, the two systems were compared directly to each other with regards to sensitivity in saliva. As shown in Table 4, Cohen’s Kappa of 0.493 suggested moderate agreement between the two systems for SARS-COV-2 detection in saliva (95% Confidence interval 0.365- 0.622). However, the result of the McNemar Test showed a significant difference in sensitivity between the two systems (p<0.0001; 95% Confidence Interval 0.001-0.165).
Table 4: Degree of Agreement between the Cepheid® and Biomeme Franklin® Systems. The two systems were compared to determine how much they agreed with each other on the test results. Cohen’s Kappa was used to determine the degree of agreement between the two systems, and the McNemar Test was utilized to determine if there was a significant difference in the sensitivity/specificity of the two systems or not.
Relationship Between Sensitivity of Each System and Days Between Suspected Onset of Disease and Date of Sample Collection for CLIA and Research Tests
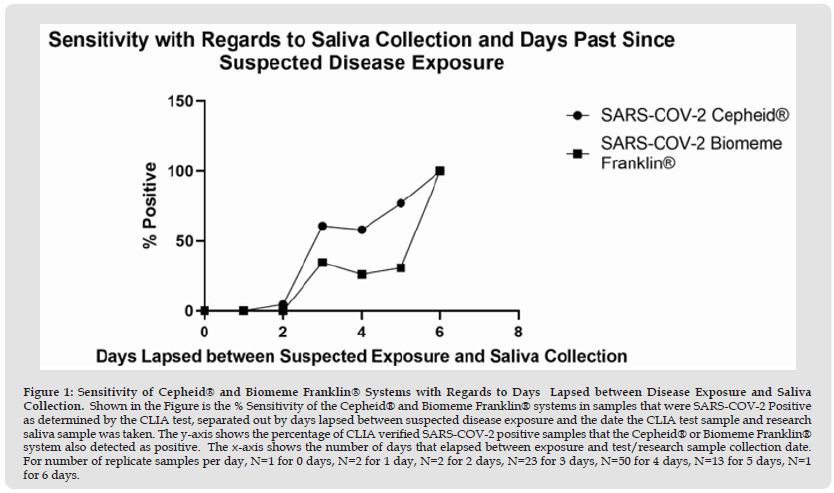
The last parameter evaluated was how the sensitivity of the two systems changed with respect to the number of days that passed between suspected exposure to SARS-COV-2 and when the actual saliva sample was collected. As shown in Figure 1, the Cepheid® GeneXpert® Xpress system demonstrated higher sensitivity than the Franklin® ISP system over the course of 0-5 days that passed between suspected exposure to SARS-COV-2 and the day of saliva collection. Only for six days that passed between suspected exposure and saliva collection did both systems show equal sensitivity. However, care should be taken with this data in regards to number of replicates. As noted in the Figure 1 legend, out of our total patient population of samples for this study, only one patient had 0 days pass between suspected exposure and saliva collection, two patients had one day pass between suspected exposure and saliva collection, two patients had two days pass between suspected exposure and saliva collection, 23 patients had three days pass between suspected exposure and saliva collection, 50 patients had four days pass between suspected exposure and saliva collection, 13 patients had five days pass between suspected exposure and saliva collection, and only one patient had six days pass between suspected exposure and saliva collection. So out of the total number of days, only three, four and five days between suspected exposure and saliva collection had more than 10 patients to analyze.
Figure 1 Sensitivity of Cepheid® and Biomeme Franklin® Systems with Regards to Days Lapsed between Disease Exposure and Saliva Collection. Shown in the Figure is the % Sensitivity of the Cepheid® and Biomeme Franklin® systems in samples that were SARS-COV-2 Positive as determined by the CLIA test, separated out by days lapsed between suspected disease exposure and the date the CLIA test sample and research saliva sample was taken. The y-axis shows the percentage of CLIA verified SARS-COV-2 positive samples that the Cepheid® or Biomeme Franklin® system also detected as positive. The x-axis shows the number of days that elapsed between exposure and test/research sample collection date. For number of replicate samples per day, N=1 for 0 days, N=2 for 1 day, N=2 for 2 days, N=23 for 3 days, N=50 for 4 days, N=13 for 5 days, N=1 for 6 days.
The purpose of this study was to evaluate and compare the sensitivity and specificity of the Xpert® Xpress CoV-2/Flu/RSV plus cartridges on the Cepheid® GeneXpert® system, and the XCEL™ Respiratory ISP cartridges on the Biomeme Franklin® ISP system in saliva. Overall, it was found that both systems exhibited high specificity for SARS-COV-2 detection in saliva. However, the Xpert® Xpress CoV-2/ Flu/RSV plus cartridges on the Cepheid® GeneXpert® system outperformed the XCEL™ Respiratory ISP cartridges on the Biomeme Franklin ® ISP system in saliva, although the sensitivity of the Cepheid® system in saliva for SARS-COV-2 detection was not very high (~56%) compared to the SARS-COV-2 CLIA test result with nasal swabs (Gold Standard). Previous studies have found saliva to be a promising sample source for SARS-COV-2 detection. For example, one meta-analysis comparing multiple studies found sensitivity in saliva to be about 88% compared to nasopharyngeal swabs [17]. Another recent study comparing saliva samples to anterior nasal and nasopharyngeal swab samples found saliva to be more sensitive than anterior nasal swabs (94.6 vs 82.6%) for the omicron variant [18].
An additional study compared saliva and midturbinate samples for SARS-COV-2 and found similar sensitivity, although saliva was more sensitive within the first few days of disease (19). Similarly, a study comparing nasal swabs to saliva for RT-PCR and digital droplet PCR also found similar sensitivity between both sample types [20]. Two previous studies compared multiple sample types including anterior nasal swabs and saliva on 1) the Biofire® RP 2.1 panel and an earlier version of the Cepheid® GeneXpert® Xpress SARS‐CoV‐2/Flu/ RSV assays [21] and 2) the Biomeme SARS-COV-2 GO-Strips on the Franklin™ three9 system and the Biofire® RP 2.1 panel [15]. It was found that saliva performed comparatively well for all systems in both studies compared to nasal swabs-95% and 98% detection on both systems respectively, in saliva compared to nasal swabs for each study [15,21].
One possible reason for the two systems in this study having much lower sensitivity in saliva compared to nasal swabs is the performance of the systems themselves. The earlier studies used separate methods for RNA extraction, cDNA synthesis and detection all on different systems [18-20]. In contrast, the two systems in the current study utilized fully integrated sample processing platforms. One of the previous studies comparing saliva to other sample types including nasal swabs used the Biomeme SARS-COV-2 GO-strips on the Franklin™ three9 system with separate RNA extraction on their M1 Sample Prep Cartridge [15], while our study used the newer XCEL™ Respiratory ISP Cartridges on the Franklin® ISP system, a fully integrated sample processing system. Thus, the separate RNA extraction step may play a role in sensitivity in saliva. However, another study compared two fully integrated sample processing systems similar to our current study and found 98% detection in saliva compared to nasal swabs for both platforms [21].
The main difference was that previous study that compared the BioFire® to the Cepheid® system used the first-generation Cepheid ® GeneXpert® Xpress SARS‐CoV‐2/Flu/RSV assay, while this study used the more recently approved second-generation Xpert® Xpress CoV-2/Flu/RSV plus assay [21]. Also, both of the previous studies utilized research samples that were collected a few days after the clinical sample was collected for the SARS-COV-2 CLIA test, so those studies were only able to compare the research samples to each other on the research systems being tested in the study. The current study directly compared research saliva samples on our two experimental systems to the clinical nasal swab sample CLIA test result.
There are a few limitations to this study. First is the saliva collection method. The previous studies [15,18-21] relied on the patient drooling into a cup to collect the saliva resulting in a whole saliva sample. The current study used a filter-based collection device in the hopes of obtaining a cleaner saliva sample that would be less prone to clogging an integrated sample processing system. The device works by the patient drooling into the sample collection tube and then the saliva passes through a filter to remove debris, impurities and reduce viscosity. However, it is possible that perhaps the filter on this device may have non-specifically bound SARS-COV-2 viral particles, thus reducing the overall amount of SARS-COV-2 viral particles in the final saliva sample for detection. On the other hand, the LoD experiments which involved spiking inactivated SARS-COV-2 virus into SARSCOV- 2 negative patient saliva that was already pre-filtered revealed that neither platform was able to detect very low copy numbers of the virus in saliva. This lends some evidence to the fact that these platforms may just not be very compatible with the saliva matrix.
A second limitation is that both systems exhibited failed runs on some of the samples. The Cepheid® Xpert® Xpress CoV-2/Flu/RSV plus assay had 1.82% of the runs fail, while the XCEL™ Respiratory ISP Cartridges on the Franklin® ISP system had 27% of the runs fail. Therefore, more refinement of the systems is required to reduce the number of failed runs if used with saliva patient samples. The last limitation is that only research saliva samples were collected, and not research saliva samples and research nasal swab samples. However, it should be noted that this study was designed to specifically evaluate these two platforms for SARS-COV-2 detection in saliva compared to the gold reference standard, rather than compare the two systems performance in saliva vs nasal swabs. This is why the current study only collected research saliva samples. By ensuring the research saliva sample was collected the exact same day as the nasal swab sample for the CLIA test, this allowed for a direct comparison to the gold reference standard-CLIA test result from a nasal swab sample, in this study.
Overall, the results of this study demonstrate while both systems in this study can detect SARS-COV-2 in saliva with high specificity, the sensitivity is not as high when compared to nasal swab samples tested by the CLIA lab. This may be due to the systems themselves, or partly due to the filter-based saliva collection device. Future studies will be needed to identify the cause and further refine/optimize the systems for SARS-COV-2 detection in saliva specifically.
We’d like to acknowledge the personnel at the Center for Advanced Molecular Detection for assisting with protocol support. We would like to thank Dr. August Blackburn for reviewing the manuscript and useful and insightful discussions.
AJB: conceptualization, experimental design, resources, funding acquisition, data analysis, manuscript drafting, editing and review. BG: data collection, manuscript review and editing. HL: data collection, manuscript review and editing. SA: supervision, resource allocation, project management, manuscript review and editing. All authors have read and approved the final manuscript.
The views expressed are those of the author(s) and do not reflect the official views or policy of the Department of Defense or its Components. The views of Cepheid® and Biomeme, Inc are not necessarily the official views of, or endorsed by, the U.S Government, the Department of Defense, or the Department of the Air Force. No Federal Endorsement of Cepheid® and Biomeme, Inc is intended.
Funding was obtained from the Defense Health Agency CARES Act. The authors have no conflicts of interest to report.
Data available on request. The data underlying this article will be shared based upon reasonable request to the corresponding author.


