Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Petra Hirjakova*, Lubomir Arnold, Marianna Jagelkova and Gabriel Minarik
Received: May 16, 2023; Published: June 19, 2023
*Corresponding author: Petra Hirjakova, Medirex group academy n.p.o., Novozamocka 67, Nitra, Slovakia
DOI: 10.26717/BJSTR.2023.51.008052
Medirex Group Academy biobank (MGA biobank) is a multidisciplinary disease-oriented project research biobank established in Nitra, Slovakia in October 2020. It evolves thanks to multiple research project and biomedical studies organized by non- profit organization Medirex Group Academy n.p.o. (Nitra, Slovakia). During 2020-2023, 27 clinics and 84 physician and nurses from 12 medical fields to participant enrolment, data and sample collecting and biobanking were involved. All together 4,346 participants were enrolled, 18,958 samples of different kind were collected and primarily processed for many downstream applications and for future use in research. Biobank is currently storing more than 53,000 biospecimen aliquots (whole blood, serum, plasma, stool, nasopharyngeal swab, native tissue, FFPE). For collecting patient data and information about samples was developed in-house MGA biobank information system (MGA BIMS). The aim of this study is to describe and evaluate current state and to define future prospective and development strategies. Since there is still no national biobank in Slovakia, it is important to further develop MGA biobank, to adapt standard processes so that in the future it meets all the requirements for implementation into European biobank structures and potentially became one of the important biobank nodes in Slovakia.
Keywords: Clinical Study; Sample Biobanking; Biospecimen Aliquots
Abbreviations: BBMRI-ERIC: Biobanking and Biomolecular Resources Research Infrastructure European Research Infrastructure Consortium; BIMS: Bioinformation Management System; DM: Diabetes Mellituse; CRF: Electronic Case Report Form; IBD: Intestinal Bowel Disease; ICF: Informed Consent Form; MGA: Medirex Group Academy
A critical requirement for personalized medicine is the availability of a large collection of patient samples with well annotated clinical and pathological data, biobanks thus play an important role [1]. Biobanks are complex systems designed for the collection, processing, storage and distribution of biospecimens and related clinical and experimental data for use in basic, translational and clinical research [2]. Considering the involvement of biospecimens obtained from human individuals, different ethical issues such as the informed consent model, sample ownership, veto rights, and biobank sustainability are debated. In connection with the collection of information and samples in the biobank, there is also a need to use efficient and robust software, which will ensure that the data will be collected in the required quality and will not be lost [3-5]. In the face of these methodological and ethical challenges, international organizations such as BBMRI-ERIC (Biobanking and Biomolecular Resources Research Infrastructure – European Research Infrastructure Consortium) Directory play a key role in supporting biobanking activities [6]. There are currently 20 EU countries listed within the BBMRI-ERIC and the highest density of biobanks is in the UK, Sweden, the Netherlands, France, Italy, and Germany [7]. Slovakia is not a member of the BBMRI-ERIC yet. It is planned that Slovakia within the BIOFORD project will become a member of this pan-European network in 2023 at the latest. However, currently, it offers a fragmented landscape of individualized biobanking organizations mainly associated with a university and general hospital units (e. g. National Institute of Rheumatic Diseases (in Slovak NURCH), National Cancer Institute (in Slovak NOU), St. Elizabeth Cancer Institute [8].
MGA biobank was founded as a local unit to support specific research projects of Medirex Group Academy n.p.o. (MGA). MGA is a non-profit organization settled in Nitra, Slovakia and focused primarily on the field of science and research, especially in biomedicine. The main goal of the projects implemented by MGA is the search for new biomarkers in order to increase preventive, diagnostic and therapeutic approaches with the potential to implement the knowledge of science and biomedical research into diagnostic and therapeutic practice. In connection with the implementation of many biomedical projects focused on different medical fields (oncology, infectology, civilization diseases), there was a need for systematic and organized sample collection. Therefore, MGA biobank was established in October 2020 as a multispecialistic academic research biobank. The systematic collection of samples continues to these days in cooperation with many health care providers from Slovakia and Czech Republic. The aim of this study is to describe and evaluate current state and to define future prospective and development strategies. From practical point of view since there is still no national biobank in Slovakia, it is important to further develop MGA biobank, to adapt standard processes so that in the future it meets all the requirements for implementation into European biobank structures and potentially became one of the important biobank nodes in Slovakia.
MGA Biobank Statute and Ethical Statement
The operation of the biobank is directed by internal statute of MGA biobank. MGA carries out activities mainly in the field of research, development, scientific, technical services and information services. The activities of the MGA biobank are managed by the Council of the MGA biobank, which has 3 (three) members from the ranks of the non-profit organization MGA. MGA is authorized to use and store samples (blood, blood components, stool, urine, saliva, isolated proteins and nucleic acids, tissue samples) for the purposes of further biomedical research, exclusively for its internal needs, which does not serve for the use of samples for persons (such as transplantation of tissue samples). The study protocols supported by MGA have been approved from the competent multicentric and local ethical committees (e. g. The ethics committee of the self-governing region of Bratislava, The ethics committee of the national oncology institute, The ethics committee of the East Slovak Oncology Institute, The ethics committee of the Trencin Faculty Hospital etc.) The prerequisite for the enrolment in the MGA studies, as well as the aims and purposes of MGA Biobank, were depicted in the written informed consent form (ICF). The main condition for the patient enrolment to the study was the signing of ICF, in which all information related to the research itself, the collection protocol, the usage and storage of samples in the MGA biobank and using of unused samples for future biomedical research were mentioned and explained by medical personnel. The sample can only be processed with the signed ICF of the participant. The type of consent depends on the study and research conditions, collection conditions, type of health care provided and subsequent processing of the sample.
MGA Biobank Standard Procedures
During all steps of biobanking process, from the participant data collecting, the sampling, the sample storage and release of biological specimens and data, we are using standardized protocols, in which are defined preanalytical conditions, primarily sample processing, documentation review steps, aliquots and derivates barcoding, storage system and container barcoding. The MGA Clinical Supervisor provides participant data quality control and MGA Biobank Supervisor prepares, supervises, updates and makes available the latest version of all protocols.
MGA Bioinformation Management System (MGA BIMS)
MGA Biobank Information Management System (MGA BIMS) is in-house developed, web-based application for the entry of data and to store of the entered data from the realised biomedical studies (i. e. participant data, sample collection data, sample processing data). Application uses a three- factor authentication log in system (username, password, grid card code) and has a modular structure consisting from two large modules divided into smaller structures (system tabs, forms, fields). One module of the system is the electronic Case report form (eCRF) serving primarily doctors and other medical personnel for the management of processes associated with study and participant administration. Another system module is intended for entering and managing samples in the biobank environment.
Software and Hardware Architecture: The hardware architecture of the application is based on an internal cluster managed by the company. Individual parts of the cluster are operated in various specific environments depending on the availability of the public Internet network. From the point of view of this availability, these environments are configured to ensure security and limit access only to absolutely necessary resources. In order to strengthen the security, evaluation of the operation and its security, the application is solved with the help of active tools for the management of network communication. The application is based on java and spring technologies and is divided into two parts. The UI part ensures rendering of graphical output for users on their web browser, using the VAADIN framework. The backend part provides services for handling requests from the UI such as persistence, data selection and processing of processes defined in the application. Individual parts of the application work within isolated containers based on docker technology and managed by the Kubernetes system or docker compose.
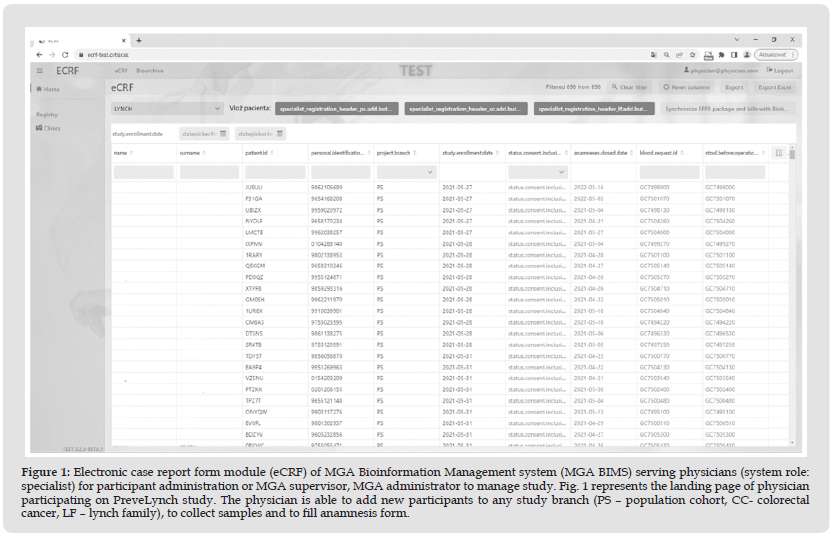
Electronic Case Report Form Module (eCRF module) Usage and Structure: The main functionality of eCRF is the collection, storage and distribution of data to users collaborating on research projects. The module covers multiple functions like complete study design, study management, reporting, participant enrolment to the specific study and participant data collection. Process of patient enrolment consist from participant registration, informed consent collection, recording, editing, saving and processing of different kind of participant data (i. e. anamnesis, laboratory results, pathology results) and collection protocol tracking. All those processes can be managed in eCRF (Figure 1), so sample collection can follow a designed collection protocol that defines work procedures. Additionally, system stores data in coded format to hide identity of patients during any research activities. To ensure bioethical safety system generates unique pseudo-anonymized IDs, which ones during registration gets every participant. Patient ID consist of 5 digits (random number and letter combination), what makes it very specific end unique for every study participant and prevents the patient from being re-enrolled in the same study or the same patient ID being assigned to another participant. For participant data collection are mostly used dynamic forms including free text, radio buttons, dropdown lists, checklists. The use of pre-defined answer choices such as those in radio buttons, checklists, and dropdown lists provides constraints during entry and, along with on-screen edit checks, are associated with higher data quality. Likewise, registration forms and forms for recording information about collected samples also contain predefined fields such as the date of enrolment the patient in the study, automatic calculation of the patient's date of birth from the birth number, date of collection to facilitate the doctor's work with the system and ensure higher data consistency.
Figure 1 Electronic case report form module (eCRF) of MGA Bioinformation Management system (MGA BIMS) serving physicians (system role: specialist) for participant administration or MGA supervisor, MGA administrator to manage study. Fig. 1 represents the landing page of physician participating on PreveLynch study. The physician is able to add new participants to any study branch (PS – population cohort, CC- colorectal cancer, LF – lynch family), to collect samples and to fill anamnesis form.
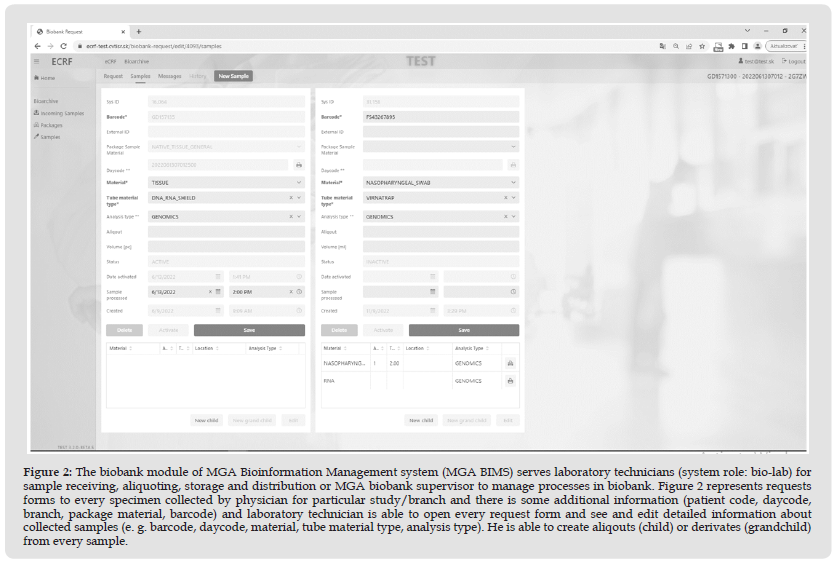
Biobank Module (Biospecimen Management Module) Usage and Structure: The module ensures the registration of incoming samples, their storage, supports the creation of derived samples of various types and their distribution to project partners. Incoming samples (mother) are entered into the system automatically based on requests from the eCRF module or manually. Derived samples can be created in two levels of the sampling hierarchy → 1st level of the derived sample hierarchy (child) → 2nd level of the derived sample hierarchy (grandchild). Samples as well as aliquots or derivates, registered in the system, can be distributed to a registered partner on the basis of a request, based on the approval of the biobank ethical committee. Distribution of samples is based on the protocol predefined in the module. System can also track current volume of available sample/aliquot/derivate considering its previous distribution (Figure 2).
Figure 2 The biobank module of MGA Bioinformation Management system (MGA BIMS) serves laboratory technicians (system role: bio-lab) for sample receiving, aliquoting, storage and distribution or MGA biobank supervisor to manage processes in biobank. Figure 2 represents requests forms to every specimen collected by physician for particular study/branch and there is some additional information (patient code, daycode, branch, package material, barcode) and laboratory technician is able to open every request form and see and edit detailed information about collected samples (e. g. barcode, daycode, material, tube material type, analysis type). He is able to create aliqouts (child) or derivates (grandchild) from every sample.
The User Management: Part of the application is the user management and their permissions to access individual parts of the application and their functionality. The permission system implements advanced options for defining access to elementary functions of the application and is divided into three hierarchical parts. The first part are the permissions themselves, which define an elementary approach to the functionality, with which they relate to. They mostly provide CRUD access, but there are also permissions for displaying part of the application or specific data. The second part are features. Because permissions are elementary, and it is necessary to group them into logical units at the level of certain functionality. This grouping facilitates the definition of the third part, where the roles themselves are defined. The user can have one role in the application and on an individual study. The permissions themselves are divided into two parts, global and contextual. Global permissions concern the application itself and are defined for everyone. Context permissions are specifically dependent on the context of the particular user. That context is study. This permission system makes it possible to define different user roles in different studies.
Database Structure: The database used in application is MariaDB, which is a relational database. The main database structures are divided into two groups. The first group ensures the definition of forms, their sections and fields used in individual studies. The second group stores input data from defined forms linked to study participants. In the case of the biobank module, the main structure is divided into tables with requests and the samples themselves. The list of distributed samples is in the protocol table, and the definition of the list of partners to whom the protocol can be distributed is defined in a separate table. Both applications use a separate database for defining users and their authorization over the applications.
Evaluation of Software Features and Performance Efficiency: Application testing takes place in two phases. The first phase is automatic testing using defined JUnit tests. This testing happens in the build phase of the application. The second phase is testing the application by users in the TEST environment. After the solution has been approved, the application is then deployed to the PROD environment. Internal application logs and profiling tools are used to ensure the speed and efficiency of the application.
Reporting/DataWareHouse/Analyses Module: System has also reported part for generating study specific reports needed for effective study management. Apart from the reporting tool in the module, a more advanced selection of specific data and their reporting using the OpenSearch tool (Amazon Web Services, Seattle, Washington, USA) is implemented in separate parts of the cluster. The OpenSearch is open-source and analytics suite, which provides a highly scalable system for providing fast access and response to large volumes of data with an integrated visualization tool, that makes it easy to explore data.
MGA Projects and Biomedical Studies
During the years 2020-2021, we started the implementation of several large-scale scientific research projects financed by the operational program Integrated Infrastructure (OPII) aimed primarily at the development of new screening and diagnostic methods in the field of oncological diseases. At the same time, in connection with the beginning of the COVID-19 pandemic, we also started participating in projects aimed at solving the issue of COVID-19. So, all samples were collected as part of these longitudinal and cross-sectional disease-focused projects, including endometrial cancer, colorectal cancer, acute COVID-19 infection and long COVID-19 syndrome, intestinal bowel disease and diabetes mellitus (Table 1). Oncology-related projects include the Biomedires II project and the PreveLynch project. The Biomedires II project is specifically focused on the issue of endometrial cancer and endometrial pathologies that have the potential to lead to endometrial cancer. The goal of the project was to find potential diagnostic, prognostic or therapeutic biomarkers detected by liquid biopsy associated with the risk of development of endometrial cancer. Another goal of the project is using of artificial intelligence for analysis the results of different laboratory methods (genomics, proteomics, glycomics) with histology and anamnesis information for more effective analysis of the detected endometrial findings. Part of the project was the realization of a biomedical study, of which the goal was to obtain clinical samples and data from patients with endometrial pathology or endometrial cancer.
The PreveLynch project aims to examine in detail the issue of hereditary cancer caused by Lynch syndrome and to investigate into the presume genetic background of the higher prevalence of selected types of cancer in the Slovak population. An important project objective is also the development of prototypes of novel, less invasive cancer screening methods based on liquid biopsy. The introduction of non-invasive screening tests involving innovative molecular methods would improve the adherence of individuals at higher risk of colorectal cancer to the established screening regimen. This should lead to higher efficiency in preventing the development of colorectal cancer or possibly other cancers associated with Lynch syndrome, resulting in an overall improvement in the health of the general population. VA-COVID-19 study supports two projects - Promedicov and DIACOVID. The both projects are focused on COVID-19 and related infectious diseases. The main goal of Promedicov project is to create a (universal) system for early and rapid detection, identification and diagnosis of new infectious diseases with pandemic potential, implemented in the pilot phase in direct connection with the current COVID-19 pandemic.
The goal of DIACOVID project is through research of serious civilizational diseases (diabetes mellitus, intestinal bowel diseases) and their complications caused by acute respiratory tract targeting viral diseases (the disease COVID-19 will serve as a model) obtain such knowledge that will enable the support and development of laboratory and clinically applicable innovative procedures for personalized diagnostics and therapy of such patients. All these projects and associated studies were designed under the professional supervision of the guarantor of the entire project with regard to the fulfilment of the set scientific goals and were approved by the relevant ethical committees. The protocol and sampling schedules for patients enrolled in these studies differed depending on the branch of the study in which they were included or according to the primary diagnosis (Table 1). The samples and data obtained in these studies were subsequently used for research by our project partners from various academic and scientific institutions in Slovakia. (e. g. Slovak Academy of Science, Geneton s.r.o., Comenius University Science Park).
Involved Medical Personnel, Patient Enrolment and Collection Protocols
Biomedires II Study Report: 67 physicians and 17 nurses from the fields of gynaecology, onco-gynaecology, gastroenterology, surgery, genetics, internal medicine, infectiology, diabetology and pulmonology cooperated with us in the recruitment of patients for the mentioned study. Patient recruitment took place in 26 clinics (hospitals and private clinics) located in various regions of Slovakia. 1,508 patients diagnosed with endometrial pathology and 268 patients with endometrial carcinoma were included until end of March 2023.
Prevelynch Study Report: The Prevelynch study included a population-based branch of 1,034 patients. Furthermore, 194 patients with colorectal cancer and 96 patients or family members of patients with Lynch syndrome were included in the study. In both cases, the collection protocol consisted of venous blood sampling for genomics and proteomics, as well as tissue sampling. Such material was subsequently sent by the physician or pathologist to the MGA Biobank, where it was primarily processed, labelled and stored according to a pre-defined protocol.
COVID-19 Related Studies (Promedicov and DIACOVID) Report: The VA-COVID-19 study was divided into two branches, within which selected subgroups of patients were followed. The first branch consisted of patients with severe civilization diseases – intestinal bowel disease (IBD), diabetes mellitus DM) who had overcome COVID-19 and control group of patients without these diseases who had overcome COVID-19. A total of 269 patients were included in this branch. The second branch was aimed at patients with an acute and varying severity of COVID-19. This branch included 431 patients with acute COVID-19, 258 patients with post-covid syndrome and a control group of 288 patients without confirmed COVID-19. Together, 977 patients were included to this branch (Table 2).
Each study had a predefined collection protocol and a set of collected anamnestic information. We tried to unify the anamnestic information so that it was as comparable as possible between the studies. The questionnaires were prepared in a modular way and particular study - specific questionnaire was compiled from the relevant modules for the given group of patients in the study. The questionnaire always included a specific set of questions related to a specific diagnosis. So, we tried to collect the same types of information in the same way for all patients.
Basic data such as height, weight, gender, age, place of residence, patient's clinical history, family history was collected from common parts across studies. A specific set of questions related to specific diagnoses of oncological diseases (endometrial cancer, colorectal cancer, Lynch syndrome), COVID-19, DM, IBD differed between the studies. For groups of patients with an oncological diagnosis, the collection protocol usually consisted of two timepoints (before and after the surgery) during which venous blood was collected for proteomic and genomic analyses, and at the same time, the collection of healthy and tumour tissue was carried out in native form and in the form of FFPE blocks by pathologist. Repeated sampling was also carried out for selected groups of patients with COVID-19. Among them, venous sampling for genomics, serology, biochemistry and immunology as well as nasopharyngeal sampling were carried out. For the other groups of patients, venous blood and/or nasopharyngeal swab sampling were performed about the relevance in the given patient group in one visit. All processes associated with patient enrolment to the study were recorded by the physician in the eCRF. Each physician/nurse had an account created in the system and was assigned study to work on. After logging into the system, the physician/nurse registered the participant by filling out the registration form, in which provided his personal data (name, surname, date of birth, ID number, residence) and confirmed that the participant had given informed consent. The system then assigned the participant a unique patient ID (pseudo-anonymous identifier), under which all his other data were recorded in the system. Only the doctor had access to the patient's personal data in the system. After giving consent, the doctor and the patient filled out the anamnestic form and entered the data on the collected samples of biological material separately according to the type of material blood/stool/tissue/smear/FFPE (barcode requests, date, time of collection) or by visit number.
Sample Collection, Transport and Pre-Analytical Requirements: The key criteria for the high-quality biobanking are to ensure the correct sample handling procedure from the sampling to storage, such as the collection and evidence of necessary data about participants for research purposes. MGA biobank include different types of biological material (blood, plasma, stool, native tissue, FFPE, swab). For the participants to be included in the study, they had to have the completed request form for each sampling material containing the basic data about patient such as name, surname, patient´s ID, date and time of collection, type of taken biological material, volume or amount of material, stamp of the clinic and doctor´s signature. The patient had to be informed about the current biomedical research and biospecimen storage in the biobank and had to give signed informed consent. These papers were archived in the administration of MGA biobank. For each type of biological material, specified conditions of the sampling, transport, processing and storage were determined, which are described in more detail below. Whole blood samples were collected in one (Promedicov) or three (Biomedires II, PreveLynch and DIACOVID) K3EDTA tubes (with the maximum volume of 10 ml for one tube) and in one serum-gel tube (with the maximum volume of 10 ml) (Promedicov). Subsequently, these specimens were always transported to the laboratory in a cool regime (2-10ºC) and in order that sample processing was ensured within 6 hours after blood collection.
The biopsy material for genomics consisted of tumour and healthy native tissue (Biomedires II, PreveLynch and DIACOVID), and FFPE tissue (Biomedires II and PreveLynch). Transport and storage of FFPE blocks was provided at room temperature. The native tissues were taken into tubes with the DNA/RNA Shield (Zymo research, California) and pre-processed in pathology lab after surgery. They were divided to two aliquots (one of tumour tissue and one of healthy tissue) and were sent in a cool regime (2-10ºC) to MGA biobank for the storage at -20°C. Stool was collected in the fecal collection tubes with DNA/RNA Shield in the volume of 10 ml from each participant (PreveLynch and DIACOVID) and was transported to the MGA laboratory, where was archived in the biobank’s freezer at -20°C until it was used for DNA isolation for microbiome analysis. Nasopharyngeal swab specimens (Promedicov) for genomic analysis were taken into the tubes containing the viRNAtrap solution for the stabilization and inactivation of infectious agents which is important for safe work in the lab. These samples were transported in cool regime (2-10ºC) to MGA biobank. After receiving the biological material in the MGA laboratory, it was activated in BIMS. For every activated sample, system generated unique daycode. All relevant information about the patient and doctor from the paper of the request form have to correspond to those in the BIMS system. During the process, the sample data (date and time of sampling, amount or volume, number of aliquots, type of analysis for which the specimen was processed etc.) were uploaded into BIMS system.
Sample Biobanking – Primary Processing, Aliquoting, Storage: Some of the specimens were treated and aliquoted directly at the pathology laboratory after the sampling (native tissue, FFPE), and the other samples were sent to the MGA laboratory for processing (blood, stool, swab). Peripheral blood was processed in two ways according to the type of analysis for which was intended. Before plasma separation, whole blood in K3EDTA tubes was taken in two aliquots of 500 μl (Biomedires II, PreveLynch, DIACOVID) or three aliquots of 1 ml (Promedicov). Subsequently, plasma for proteomic analysis was processed by single step centrifugation (2,200 g, 10 min., 4°C) and was divided into two aliquots of 500 μl, and further aliquots of 1,8 ml. Plasma for genomic analysis was separated by two-step centrifugation (2,200 g, 10 min., 4°C; 16,000 g, 10 min., 20°C), and was divided into two aliquots of 680 μl, and further aliquots of 1,8 ml. Individual blood and plasma aliquots was labelled by their daycode separately for proteomics and genomics. Blood collected into serum-gel tubes for serology was processed by centrifugation (2,200 g, 10 min., 4°C) and aliquoted into two serum aliquots of 1 ml.
As regards FFPE blocks and the native tissues, it was not necessary to aliquot them. They were archived in the biobank in the form in which they were delivered. Stool was aliquoted in the volume of 1 ml (one aliquot of each specimen) and swab originated solution in the volume of 1.5 ml (two aliquots of each sample). All samples were labelled with the corresponding daycode immediately after activation in the BIMS system. All specimens except swab originated solutions and FFPEs were stored in the box marked with a label that contained the information, such as the type of analysis, the sampling material, the daycode of the first sample which is placed in the box, and were archived in chronological order at -20°C. FFPE material of tumour and healthy tissue, also marked with the corresponding daycode, was kept at room temperature (one piece of each). Swab originated solutions were stowed in the box labelled with the daycode of the first and last sample at a temperature of 2-10 °C. The storage of samples in the biobank were divided into the separate freezers and refrigerators according to the study.
Sample Cycle in Biobank: The sample cycle of biobanking starts from collection, processing, storage, maintenance, distribution, utilization, evaluation of analysis and ends the registration of the results. The specimens are archived until they used for further analysis and testing. The key point for the sample distribution from the biobank have been the signed official request that had to contain the identification of the requester, the project title, the short annotation of the study, the objectives of the study, the method description, the list of workers authorized to the work with these samples and as the attachment – the protocol for release of certain type of samples containing the exact list of required samples. After submitting this request and its approving by the Council of the MGA biobank, it has been necessary to create and to register the mentioned protocol containing the list of required samples from the selected study in the BIMS system. The printed version of the protocol had to be signed by the MGA Biobank supervisor and the partner´s responsible person together with signed request, both have been archived in the administration of MGA biobank. Distributed samples can be used for the extraction of DNA/RNA (e. g. human, bacterial, viral), proteins, glycans for the purpose of research focused on genomic, proteomic and glycomic analyses, analysis of microbiome, of which the results can be shared between the individual research project workplaces.
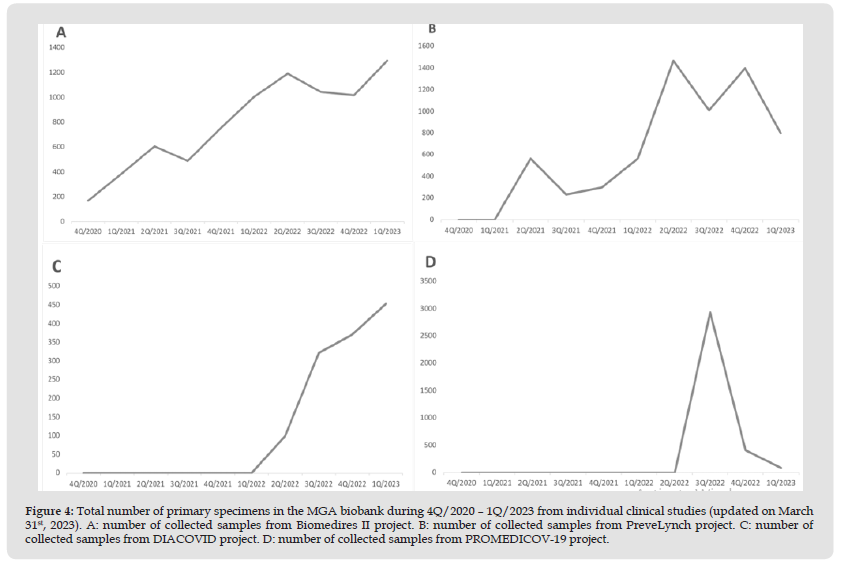
Biobanking – Current Stage: Currently (March 31st, 2023), the MGA biobank is a repository of 18,958 different biological samples in totally 53,600 aliquots from 4,346 patients participating on four main projects focused on oncological (PreveLynch, Biomedires II) and infectious (DIACOVID, Promedicov) diseases (Table 3). The first samples have been collected in November 2020 (Biomedires II study), in April 2021 (Promedicov), in May 2021 (PreveLynch) and in June 2022 (DIACOVID). Number of samples received and stored in the MGA biobank in the individual quarters of the year during 2020-2023 is graphically illustrated in (Figures 3 & 4). There are currently stored 13,523 blood samples, 1,949 FFPEs, 1,544 stool specimens, 1,190 SWAB originated solutions and 752 native tissues. The biobanking of samples continues throughout the entire duration of individual projects. Archived biological material is gradually used for various analysis types (genomics, proteomics, glycomics, serology, immunology, histology). To date, more than 2,500 aliquots have been released to participating partner´s organizations (Slovak Academy of Science, Comenius University Science Park, Medirex Group Academy Bratislava, Geneton s. r. o.) for genomic analysis from swab originated solutions and the plasma (VA-COVID-19 study), from the whole blood, plasma, the tumour and healthy native tissue (Biomedires II), from the whole blood and plasma (PreveLynch), and also for proteomic analysis from the plasma (PreveLynch and Biomedires II). The results of mentioned analyses are still in processing phase.
Datawarehouse: Our data analysis warehouse is currently in the process of development, but as a temporary solution for specific searching in the clinical data database, we use the commercially available analysis tool OpenSearch, which is able to perform search related jobs in data of various kinds about patients included in MGA studies from the eCRF system in pseudo-anonymized manner, e. g. anamnestic data, results of laboratory examinations, pathology results, data on collected biological samples. OpenSearch enables not only browsing through data but also creating user defined reports (dashboards) and basic data analysis and preparation of graphical outputs.
Figure 3 Total number of primary specimens in the MGA biobank during 4Q/2020 – 1Q/2023 from all clinical studies (updated on March 31st, 2023).
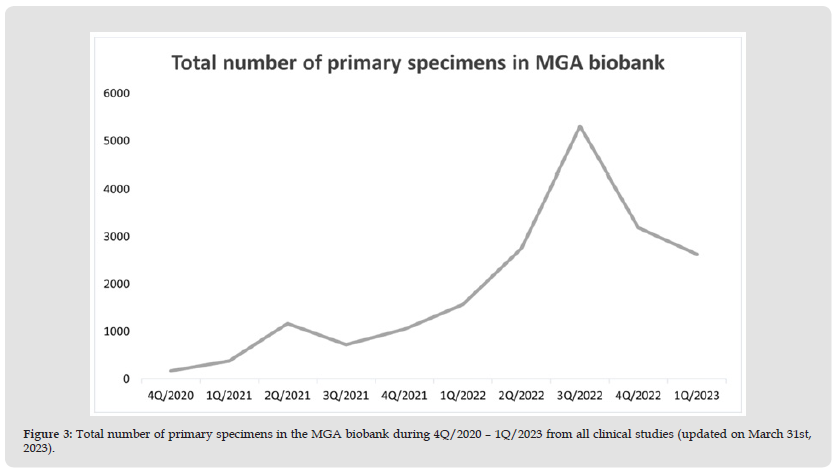
Figure 4 Total number of primary specimens in the MGA biobank during 4Q/2020 – 1Q/2023 from individual clinical studies (updated on March 31st, 2023). A: number of collected samples from Biomedires II project. B: number of collected samples from PreveLynch project. C: number of collected samples from DIACOVID project. D: number of collected samples from PROMEDICOV-19 project.
Biobanks enable wide-scale research by professional collection of biological specimens and correlated clinical data which may include health records, family history, lifestyle and genetic information that are held predominantly for use in health and biomedical research [4,5] according to requirements related to ethical and legal issues or the standardization of different processes involved in sample collection. MGA biobank was founded as a local unit to support specific research projects, therefore many processes associated with the collection of data and biological samples need to be optimized, improved and harmonized so that they are in accordance with the requirements the International Organization for Standardization for biobanking (ISO 20387:2018) [2], providing high- quality bio-specimen and data for the future research. One of MGA biobank goals is also to improve BIMS software for data collection, sample processing, storage, distribution and especially for the future, also use data warehouse for data analysis to make working with the data more efficient and easier for physicians, laboratory personnel and researchers. With such perspective and motivation in the future. MGA biobank will be prepared technically for joining the network of biobanks in the countries of the European Union and become a member of BBMRI - ERIC network, which enables to cooperate widely with foreign partners. At the same time, we plan to continue the realization of biomedical studies focused primarily on the field of civilization and oncological diseases in order to develop and support cooperation between the health sector (hospitals, clinics) and the academic sector (universities, scientific research institutions) in Slovakia. Up to now, MGA Biobank efficiently supported research in Slovakia with more than 2,500 samples released to participating partner´s organizations in the past year, which supported studies with results recently published [9,10].
This article was created with the support of the OP Integrated Infrastructure for the project: Research on COVID-19 progressive diagnostic methods and biomarkers useful in early detection of individuals at increased risk of severe disease, ITMS: 313011ATA2, co-financed by the European Regional Development Fund.


