Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Zhanpan Zhang1*, Dipen P Sangurdekar1, Susan Baker2 and Cristina A Tan Hehir1
Received: April 13, 2023; Published: April 19, 2023
*Corresponding author: Zhanpan Zhang, GE Global Research, Niskayuna, NY, USA
DOI: 10.26717/BJSTR.2023.49.007866
Blood-based protein biomarkers predicting brain amyloid burden would have great utility for the enrichment of Alzheimer’s Disease (AD) clinical trials, including large-scale prevention trials. In this paper, we adopt data fusion to combine multiple high dimensional data sets upon which classification models are developed to predict amyloid burden as well as the clinical diagnosis. Specifically, non-parametric techniques are used to pre-select variables, and random forest and multinomial logistic regression techniques with LASSO penalty are performed to build classification models. We apply the proposed data fusion framework to the AIBL imaging cohort and demonstrate improvement of the clinical status classification accuracy. Furthermore, variable importance is evaluated to discover potential novel biomarkers associated with AD.
Keywords: Alzheimer’s Disease; Data Fusion; Classification; Variable Selection
Abbreviations: PET: Positron Emission Tomography; CF: Cerebrospinal Fluid; PIB: Pittsburgh Compound B; MCI: Mild Cognitive Impairment; KNN: K-Nearest-Neighbor; SAM: Significance Analysis of Microarrays; OOB: Out-Of-Bag; AIBL: Lifestyle Flagship Study of Aging; SURR: Standardized Uptake Value Ratio; AD: Alzheimer’s Disease
Alzheimer’s disease (AD) is the most common form of dementia in later life, affecting 1 in 8 people by the age of 65 years. The diagnosis of AD can only be confirmed, with certainty, by histologic examination of the brain tissue at autopsy. A key pathological hallmark of AD is the deposition of amyloid-β (Aβ) in the brain, and there is a strong association between brain amyloid burden and the risk of developing AD-like pathology. It is believed that Aβ accumulation precedes clinical presentation of cognitive impairment by many years [1], enabling detection of preclinical AD and promoting pre-symptomatic treatment of AD should a disease modifying treatment becomes available. In living patients, Aβ burden is determined either by cerebrospinal fluid (CSF) biomarkers or positron emission tomography (PET) with Aβ radiopharmaceuticals such as 11C-Pittsburgh compound B (PiB) [2]. Recent FDA approval of longer-lived 18F amyloid imaging radiopharmaceuticals could promote their use in clinical practice [3]. Amyloid PET scan allows a semi-quantitative in vivo assessment of Aβ deposition in the subject brains because its uptake in AD correlates with Aβ plaques measured neuropathologically in the same brains [4]. However, this approach is costly and is restricted to specialized centers. CSF sampling is invasive and there are no standardized methods to handle and analyze CSF biomarkers resulting in variability across different labs [5]. For cost-effective, simple, and non- invasive testing, blood-based biomarkers that predict brain amyloid burden would have great utility in identifying subjects at risk for AD.
Previous studies have demonstrated that blood-based metabolites and autoantibodies have the potential to predict diagnosis of AD. The group at VTT identified signatures of lipids and small polar metabolites in plasma associated with the progression of mild cognitive impairment (MCI) to AD [6]. Using high-throughput antigen microarray from Life Technologies, a panel of IgG autoantibodies from human serum was shown to differentiate AD and MCI from healthy control [7,8]. The purpose of this study is to evaluate these metabolomics and autoantibody variables as well as to discover potential novel biomarkers associated with AD using an independent cohort. It is important to simultaneously analyze different types of data sets specifically if the different kind of biological variables are measured on the same samples. Such an analysis enables a real understanding on the relationships between these different types of variables. Data fusion [9-11] refers to the combination of data originating from multiple sources and is used to improve decision tasks – such as classification, estimation, and prediction – and to provide a better understanding of the phenomena under consideration. The purpose of fusion is to optimize the total information content from multiple sources. [12] pointed out that total information content can be enhanced in the case of multiple sensors fusion because new sensors can be used to provide more data, and similar sensors can be added to provide more coverage or more confidence for observed data.
Class prediction with high-dimensional features is an important problem and has received a lot of attention in biological and medical studies. The task is to classify and predict the diagnostic category of a sample on the basis of its feature profile, which is challenging because there are usually a large number of features and a relatively small number of samples, and it is also important to identify which features contribute most to the classification. Our interests lie in integrating multiple high dimensional data sets and perform variable selection simultaneously. Some sparse associated integrative approaches have been applied to include a built-in selection procedure for feature selection in integration studies. The work presented in this paper proposes a framework for improving diagnosis of AD by data fusion and model fusion. Section 2 describes the methodologies that are used to pre-process data including missing value imputation and variable pre-selection, develop and assess classification model, and evaluate the variable importance. Section 3 demonstrates how the proposed framework works for the diagnosis of AD by combining both metabolome and IgG/IgM autoantibody variables, and conclusion and discussion are included in Section 4.

Data Imputation
Missing values are imputed via the K-Nearest-Neighbor (KNN) algorithm [13]. For each target variable having at least one missing value, the nearest neighbor variables are identified which have the smallest Euclidean distance than the others. The missing feature values in the target variable are imputed by using the averages of the non-missing entries from the nearest neighbors. As a large number of variables causes much intense nearest-neighbor computations, the KNN imputation algorithm is combined with a recursive two-means clustering procedure, which recursively divide the variables into two smaller homogeneous groups till all groups have less than a specific number of variables, and the KNN imputation is performed separately within each variable group.
Variable Pre-selection
To avoid simultaneously using tens of thousands of variables and adding too much noise into the classification model development, methods are needed to pre-select a subset of more important variables. Significance Analysis of Microarrays (SAM) [14] has been widely used to determine the significance of gene expression changes between different biological states while accounting for the enormous number of gens. For a two-class response variable, i.e. C = 2 , both the t-statistic and Mann-Whitney-Wilcoxon statistic can be used to compute the score for each variable. A threshold is selected to ensure a specific False Discovery Rate (FDR) is achieved. Kruskal-Wallis test is an extension of Mann-Whitney-Wilcoxon test when there are more than two classes, i.e., C > 2 , which tests whether the feature values are from the same distribution or not. The p-values for testing all the variables are ordered, and those corresponding to the smallest p-values are considered as the most important values.
Random Forest for Classification
Random Forest [15] is a non-parametric approach that builds a large collection of de-correlated decision trees on bootstrapped samples and then averages them. Each time a split in a tree is considered, a random sample of m(m < p) variables is chosen as split candidates from the full set of p variables. For classification, a random forest obtains a class vote from each tree, and then classifies using majority vote. It is often useful to learn the relative importance or contribution of each variable in predicting the response. For each tree, the prediction error on the out-of-bag (OOB) samples is recorded. Then for a given variable Xj , the OOB samples are randomly permuted in Xj and the prediction error is recorded. The variable importance for Xj is defined as the difference between the perturbed and unperturbed error rate, averaged over all trees.
Logistic Regression for Classification
The multinomial logistic regression model is specified in terms of C −1 logit transformation:

When N < P , the L1 LASSO (Least Absolute Shrinkage and Selection Operator) penalty [16] can be used for variable selection and shrinkage, which forces some of the coefficient estimates to be exactly equal to zero when the tuning parameter λ is sufficiently large. The optimal tuning parameter λ is chosen such that the crossvalidation error is minimized. For C > 2 , a grouped-LASSO penalty on all the coefficients for a particular variable is used, which makes them all be zero or nonzero together.
Cohort
Subjects included in this study were a subset from the Australian Imaging, Biomarker and Lifestyle Flagship Study of Aging (AIBL), which is a prospective, longitudinal study of aging, neuroimaging, biomarkers, lifestyle, and clinical and neuropsychological analysis, with a focus on early detection and lifestyle intervention. The dual center study recruits patients with an AD diagnosis, MCI, and healthy volunteers with the aim of identifying factors that lead to subsequent AD development. Additional specifics regarding subject recruitment, diagnosis, study design, details of blood collection and sample preparation have been previously described [17]. The PiB amyloid PET imaging methods have been previously reported [18].
Blood Biomarker Measurements
For metabolomic analysis, plasma samples were provided to VTT Technical Research Center of Finland (VTT). Methods for global lipidomics and global profiling of small polar metabolites have been previously described by VTT [6]. To identify autoantibody signatures, serum and plasma samples were provided to Life Technologies (Invitrogen) for the ProtoArray Immune Response Biomarker Profiling Service. The array contains over 9,000 unique human protein antigens [19]. Serum samples were used for detection of IgG autoantibodies to compare with a previously published report [7]. Plasma samples from the same patients were used for detection of IgM autoantibodies.
A set of VTT metabolomic variables are measured on 197 selected subjects, including 711 Polar metabolite variables and 790 Lipid variables. A set of autoantibody variables are measured on 242 selected subjects, including 9480 IgG variables and 9480 IgM variables. Merging the above two sets of variables leads to a master data set which includes 180 subjects (116 Healthy, 43 MCI and 21 AD) and 20461 variables. A standardized uptake value ratio (SUVR) cutoff of 1.5 is used for the PiB-PET scans to divide subjects into two groups: PiB negative (PiB- with PiB SUVR<1.5) and PiB positive (PiB+ with PiB SUVR>1.5) with its distribution shown in (Table 1), where the genotype is defined as the Apolipoprotein E4 (APOE4) carrier status (E4- and E4+). Presence of the E4 allele has been identified to be a risk factor associated with AD [20]. The demographic variable summary is shown in (Table 2 & Figure 1). Log10 transformation is performed to the feature matrix, and the variables with only unique value are excluded. As the number of variables is large, the KNN imputation algorithm is combined with a recursive two-means clustering procedure, where and a maximum group size of 1500 variables is used.
PiB SUVR Classification Model
The Mann-Whitney-Wilcoxon statistic is used in the Significance Analysis of Microarrays (SAM), and the 200 most significant variables are pre-selected which ensures around 10% FDR.
The following three set of variables are used to develop the PiB SUVR classification model:
• Model 1: Age + Gender
• Model 2: Age + Gender + Genotype
• Model 3: Age + Gender + Genotype + Blood-based variables
Model 2 is built as a baseline as the demographic variables are usually available in reality. As shown in Table 1, most E4- subjects are in the PiB- group and most E4+ subjects are in the PiB+ group, therefore it is of interest to investigate the impact of genotype by only including age and gender in Model 1. Also, to study the additional contribution from the blood-based variables, all the three demographical variables are forced to be included in Model 3. Model 1 and Model 2 are built via both random forest and binomial logistic regression. When using random forest, 2000 trees are generated, the minimum terminal node size is 1, and all the demographical variables are used as candidate variables for each node split. Model 3 is built via both random forest and binomial logistic regression with L1 LASSO penalty. When using random forest, 2000 trees are generated, the minimum terminal node size is 1, and 15 variables are used as candidate variables for each node split. Furthermore, cross-validation is used for the model building and model assessment. At each time, the 180 subjects are divided into a training set with subjects and a test set with subjects. The model that is built upon the training set is applied to the test set to assess the classification performance. The above process is repeated 100 times, and the metrics of specificity, sensitivity, AUC are recorded accordingly. (Figures 2a & 2b) summarize the classification performance for random forest and binomial logistic regression, respectively, and the median metrics are summarized in (Table 3). These performance metrics show that the E4 genotype significantly improves the PiB SUVR classification performance, especially for the random forest model. However, the performance metrics for Model 3 are not quite different from those for Model 2, which implies that the blood-based variables do not significantly improve the PiB SUVR classification performance.
Clinical Status Classification Model
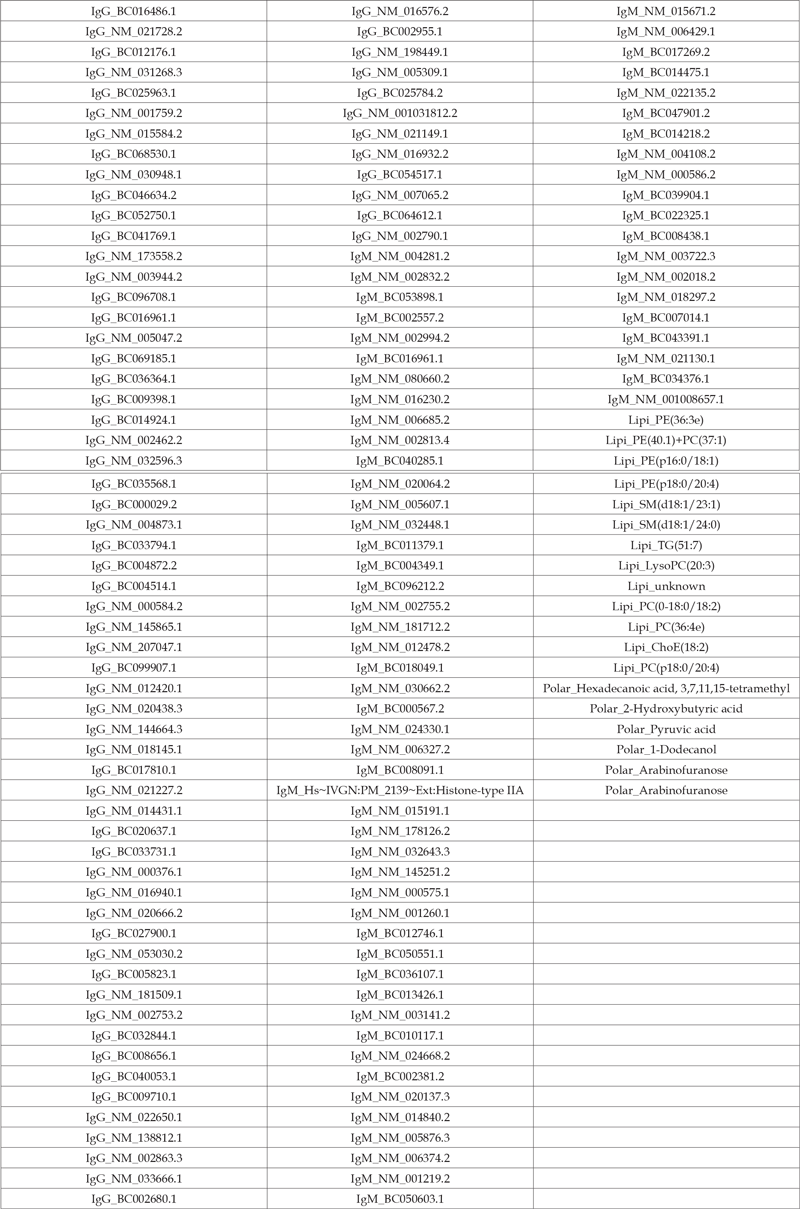
Kruskal-Wallis test is applied to each variable, and the 200 variables with the smallest p-value are pre-selected. As in Section 3.2, the same three sets of variables are used to develop the clinical status classification model. Model 1 and 2 are built via both random forest and multinomial logistic regression. Model 3 is built via both random forest and multinomial logistic regression with L1 LASSO penalty. Also, the same parameters are set for random forest as in Section 3.2. (Figures 3a & 3b) summarize the classification accuracy for the random forest and multinomial logistic regression model, respectively, and the median accuracies are shown in (Table 4). Figure 3a shows the bloodbased variables improve the clinical status classification accuracy by 12% when the random forest technique is adopted resulting in an accuracy of 66%. Figure 3b shows the classification accuracy is also slightly improved by including the blood-based variables when the multinomial logistic regression technique is adopted, but the improvement is not as much as that for the random forest technique, which is because the multinomial logistic regression model has better classification performance than the random forest model when the clinical status classification model is built upon only the demographic variables. The top variables are shown in (Table 5), most of which are IgG and IgM autoantibodies. The accuracy in our study is lower than a previous report with IgG autoantibodies [7] where it was reported to be over 90%. This disagreement could be due to differences in cohort size and composition as well as the demographic variables (balance of age and gender between the clinical classifications).
Table 5: Median metrics for clinical status classification performance.Top blood-based variables for clinical status classification.
Data fusion exists in many fields of study and can be used to create composite knowledge signatures from multiple sources by creating new signatures and improving the existing ones from raw data, adding additional signatures to the existing ones to increase coverage, and studying the dissimilarity among signatures and creating signatures that complement each other. In this paper, we adopt data fusion to combine multiple high dimensional data sets upon which classification models are developed to predict amyloid burden as well as the clinical diagnosis. Specifically, non-parametric techniques are used to pre-select variables, and random forest and multinomial logistic regression techniques with LASSO penalty are performed to build classification models. We apply the proposed data fusion framework to the AIBL cohort and demonstrate improvement of the clinical status classification accuracy. Class prediction with highdimensional features is an important problem and has received a lot of attention in the biological and medical studies [21-24]. Variable and feature selection have become research focus when tens or hundreds of thousands of variables are available. More classification modeling and variable selection techniques will be investigated in future work. Furthermore, we will consider expanding the current data fusion framework to include more data sources of different platforms.


