Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Papastefanopoulou V1,2, Simoudis A1, Charoniti E1, Papadopoulou M1, Stanitsa E3,4, Papatriantafyllou I5, Papageorgiou S3,4 and Kroupis C1*
Received: April 07, 2023; Published: April 27, 2023
*Corresponding author: Associate Professor Christos Kroupis, Department of Clinical Biochemistry and Molecular Diagnostics, Attikon University General Hospital, Medical School, National and Kapodistrian University of Athens, Greece
DOI: 10.26717/BJSTR.2023.49.007855
Introduction: Age-related macular degeneration (AMD) is a degenerative eye disease leading to significant loss of central vision. A strong association with the disease has been identified worldwide for two common coding single nucleotide polymorphisms (SNPs): A69S (rs10490924) of the ARMS2 gene involved in oxidative stress-dependent damage to the retinal photoreceptors and Y402H (rs1061170) of the complement factor H (CFH) gene involved in innate immunity. These two SNPs explain about 25% of cases of AMD. In recent literature, a link between AMD and Alzheimer’s disease (AD) has been postulated. Indeed, the possibility of early diagnosis of cognitive degeneration of the brain in AD is suggested by using non-invasive optical coherence tomography (OCT) in the retina. We aimed the investigation of the possible association between AMD and AD, as attributed by the analysis of the two aforementioned SNPs.
Materials and Methods: Peripheral blood was collected from consecutive well-ascertained AD patients of our University Hospital and their caregivers after obtaining their informed signed consent. The patients underwent a complete neurological and neuropsychological examination. In total, thirty-eight healthy subjects and 120 AD patients participated. After DNA isolation, CFH genotyping was performed by realtime qPCR-melting curve analysis developed by our research team on the LightCycler platform (Roche) and ARMS2 genotyping by a PCR-RFLP method. Statistical analysis was performed by using the SNPstats online tool.
Results and Conclusions: There was no significant correlation with either CFH [Odds Ratio = 0.90 (95% CI = 0.54-1.49), p = 0.68] or ARMS2 gene [OR=1.29 (0.60-2.77), p=0.5)]. However, this pilot study should be further extended to sub-groups of the two diseases and other genes involved in common mechanisms of pathogenesis in both disorders.
Keywords: AMD; ARMS2; CFH; SNPs; Alzheimer’s Disease
Abbreviations: AD: Alzheimer’s Disease; AMD: Age-Related Macular Degeneration; CFH Complement Factor H; CI: Confidence Interval; MCI: Mild Cognitive Impairment; OCT: Optical Coherence Tomography; OCTA: Optical Coherence Tomography Angiography; OR: Odds Ratio; PCR Polymerase Chain Reaction; RFLP: Restriction Fragment Length Polymorphism; SNP: Single-Nucleotide Polymorphism
The most common macular condition is called Age-related Macular Degeneration (AMD). It is a neurodegenerative disease that leads to loss of central vision since the affected areas are the main areas of the retina and choroid of the eye. In AMD, there is a disturbance in degradation processes, ultimately leading to extracellular deposits called drusen and to the accumulation of photoreceptors’ material (lipofuscin) in the retina pigment epithelium, significantly affecting the normal function of the overlying photoreceptors and thus vision. [1] According to epidemiological studies, age is particularly implicated in disease occurrence, with a dramatic increase for patients after the age of 80 years. Age, obesity, atherosclerosis, hypertension, and heredity are additionally important predisposing factors in developing the disease. [2] Furthermore, recent studies have identified genes significantly associated with the disease and increased risk of developing AMD. [3] The most critical representative genes are CFH [4-6] and ARMS2. [7-9] Additionally, single nucleotide polymorphism (SNP) analysis has become a valuable and attractive tool in investigating the genetic basis of multifactorial and complex diseases such as AMD. Data from studies support that polymorphism in these two genes and others encoding complement factors (C2, C3, RFB, etc.) may explain about 75% of cases of AMD [10]. The genetic basis of the disease is an essential factor for understanding its pathogenesis and possibly creating a therapeutic model.
Specifically, a strong association with the disease has been established in the Caucasian race for the A69S (rs10490924) SNP of the ARMS2 gene, as this polymorphism is involved in oxidative stress-dependent retinal photoreceptor damage. An association with disease has also been found for the Y402H (rs1061170) SNP in the complement factor H (CFH) gene involved in the alternative complement pathway regulation in innate immunity. These two common coding SNPs explain about 25% of cases of AMD [11-12]. Recently, the relationship between AMD and Alzheimer’s disease (AD) has been extensively postulated. Since CFH and ARMS2 also affect brain structure and function, they can be considered risk genes favourable for AD development. [13] Alzheimer’s disease (AD) and AMD are degenerative diseases affecting people of advanced age. They are associated with activating several common mechanisms, such as the formation and accumulation of extracellular protein and the presence of beta-amyloid and apolipoprotein (APOE) elements that constitute a significant component of the amyloid plaques configuration in AD but also in the drusen of AMD. [14] Other similarities in the pathophysiology of AMD and AD include neuroinflammation, increased oxidative stress, mitochondrial and lysosome dysfunction, and complement activation. These similarities provide a sufficient basis for many to characterise AMD as «Alzheimer’s disease of the eye.» [15].
Moreover, changes in retinal structure, metabolic state, and neuroinflammation are also involved in AD and mild cognitive impairment (MCI). [16] According to histopathological autopsies in AD patients, loss of the retina ganglion cells and a decrease in the volume of the macular layer have been observed. High-resolution noninvasive imaging tools such as optical coherence tomography (OCT) and optical coherence tomography angiography (OCTA) can detect microvascular alterations and neuronal loss in AD and, therefore, be considered biomarkers for the diagnosis and the risk of disease progression [17]. This pilot study aims to investigate the possible association between AMD and AD in a Greek cohort, as revealed by analysing the two aforementioned SNPs.
Patients
120 well-ascertained Greek Caucasian patients with Alzheimer’s disease (AD), with an average age of 72 years, were referred to the Cognitive Disorders/Dementia Unit of the 2nd Neurological Clinic of the National and Kapodistrian University of Athens at Attikon University General Hospital for a thorough clinical evaluation. Additionally, 38 healthy sex-matched individuals with an average age of 57 years, volunteers or caregivers to the patients, with no allegations of cognitive impairment, were included in the study. Medical and family history, neurological evaluation, magnetic resonance imaging (MRI) or computed tomography of the brain, and neuropsychological assessment, including tests such as Mini-Mental State Examination (MMSE) [18] and Frontal Assessment Battery (FAB) [19] were incorporated for the diagnostic procedure. AD was diagnosed respecting the McKhann criteria [20] with a CDR score of 1. The recruitment period began in 2015 and ended in 2017. All diagnoses were made by the same neurologist [SGP] with expertise in dementia and cognitive disorders. Following the approval of the local Bioethics Committee (987/9-9-15) and in accord with the General Data Protection Regulation [21], a signed consignment was handed to both groups before they participated in the study.
Genomic DNA Isolation and Genotyping
DNA isolation was performed from blood collected in EDTA tubes with a High Pure PCR Template Kit (Roche) in the Clinical Biochemistry Laboratory of the Attikon University General Hospital. CFH genotyping for coding SNP rs1061170, (GRCh38, chr1-196690107 C>T, NG_007259.1 g.43097C>T, NM_000186.4 c.1204C>T), that results in the missense substitution of histidine to a tyrosine amino acid at position 402 of the CFH protein (p.Y402H) was performed by our novel real-time qPCR-melting curve analysis developed by our research group on the LightCycler platform (Roche) as described previously [22] ARMS2 genotyping for coding SNP rs10490924 (GRCh38, chr10-122454932 G>T, NG_011725.1 g.5270G>T, NM_001099667.3 c.205G>T), that results in the missense substitution of alanine to a serine amino acid at position 69 of the ARMS2 protein (p.A69S) was performed by applying a literaturebased PCR-RFLP method with PvuII restriction [23-24]. Additionally, in all samples, APOE genotyping was performed with the LightMix APOE C112R R158C kit by TIB MolBiol in the LightCycler platform (Roche) as described in Papastefanopoulou, et al. [25] in order to detect the E4 risk allele for Alzheimer’s Disease (GRCh38, rs429358, chr19- 44908684 T>C, NG_007084.2 g7903T>C, NM_000041.4 c.388T>C) that results in the missense substitution of cysteine to an arginine amino acid at position 130 of the APOE protein (p. C130R or 112 of the secreted protein) for the development of AD disease.
Statistical Analysis
The SNPstats software through the SNPstats Internet platform (http:// bioinfo.iconcologia.net/SNPstats web) was used to examine the frequency of ARMS2, CFH, and APOE4 variants and their association with AD. For our sample, we followed the crude analysis followed by interaction analysis with sex as a covariate. The genotype distribution for ARMS2, CFH, and the APOE4 variants was checked with the Hardy-Weinberg equilibrium (HWE), and odds ratio (OR) and corresponding confidence intervals (CI) were calculated for all four inheritance models (dominant, co-dominant, recessive and logadditive). All significance tests were two-sided, and p-values <0.05 were considered significant.
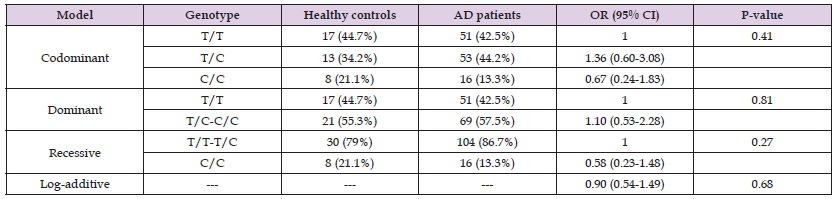
As expected, the mean age for the control group was 57.39 years and 72.9 years for patients. Sex was matched in both groups (49% females, 51% males in the AD group, χ2 test, p>0.05). All DNA samples were appropriately genotyped for the two SNPs in CFH and ARMS2 genes (characteristic images are shown in (Figures 1 & 2)) and the risk allele of the APOE gene. All genotype frequencies were in Hardy Weinberg equilibrium (p>0.05). The patient AD group was well ascertained since the E4 risk allele carriers were significantly increased in the AD group compared to healthy controls as expected [27.5% versus 13.5% in controls, p=0.027, OR 2.48 (1.21-6.18), data not shown]. The ARMS2 genotype distribution in the studied healthy and patients group associated with the disease is shown in (Table 1). The G/G homozygosity of the wild-type variant was the most prevalent, with a 73.7% and 70% rate for the control and patients group, respectively. The T/T homozygosity was found in none of the control individuals but at a 2.6% rate in the patient group. Despite this increased tendency, it doesn’t yet reach statistical significance, with the log-additive model [p=0.5, OR 1.29 (0.60- 2.77)]. Additionally, no significant association was determined when examining sex interaction (p= 0.73, (Table 2)). As shown in (Table 3), the homozygosity for the wild-type allele (T/T) of the CFH gene is counted at 44.7% for the control group and 42.5% for the group of patients. Meanwhile, the homozygosity for the C risk is 13.3% for AD individuals and 21.1% for healthy individuals. The log-additive inheritance model for the C mutant allele calculates an OR of 0.90 (95% CI 0.54-1.49, p=0.68). When stratified by gender, there is no statistical significance (p= 0.63, (Table 4)).
Figure 1 Melting curve analysis after a real-time PCR CFH rs1061170 SNP genotyping assay in the Light Cycler (Roche) platform: a mutant homozygote CC (red curve), a heterozygote CT (green curve), and a wild-type TT sample (grey curve) are shown. (Adapted from [22]).
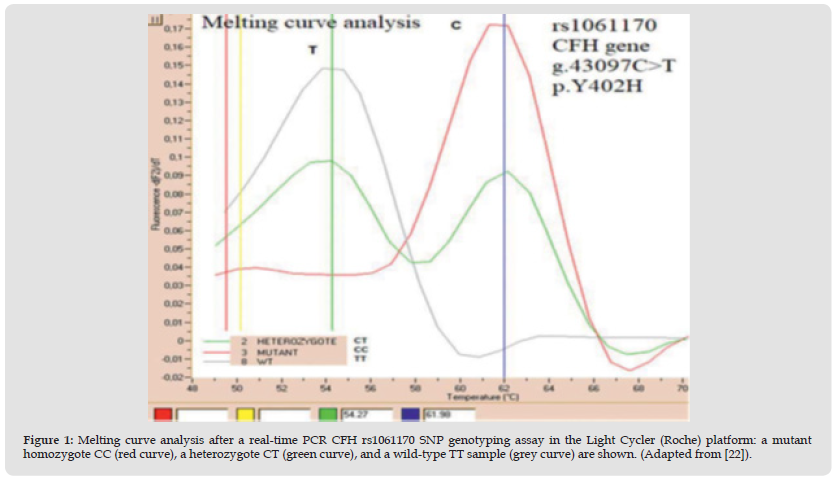
Figure 2 Agarose electrophoresis result of the ARMS2 rs10490924 SNP PCR-RFLP genotyping method. In the first two wells, the molecular weight (MW) marker (PCR Marker, NEB) and the PCR blank are shown. Wells 1,3 show homozygote mutant TT samples, well 2 a wild-type GG sample and wells 4,5 heterozygous G/T samples. (Adapted from [23]).
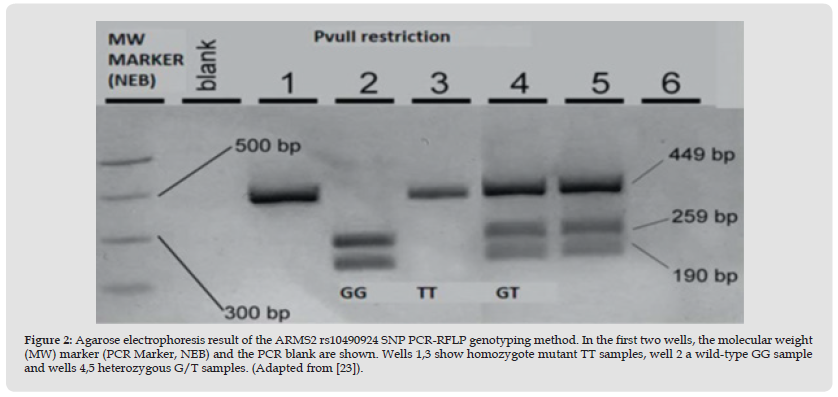
Table 1: Genotype frequencies, inheritance models, and ORs for the SNP in the ARMS2 gene (G wild-type allele and T mutant allele).
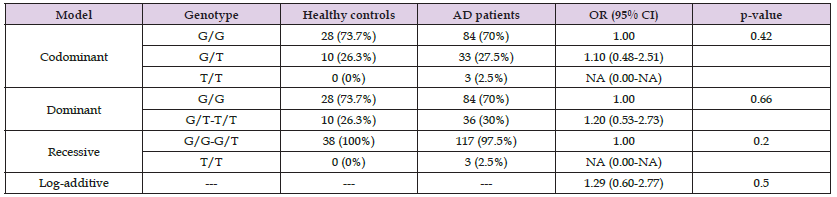
Table 3: Genotype frequencies, inheritance models, and ORs for the CFH gene SNP(T polymorphism wild-type allele and C polymorphism mutant allele).
AMD and AD are both associated with complex biological pathways regarding aging, lipid transport and metabolism, inflammation response, extracellular matrix remodelling, oxidative stress, mitochondrial damage, and alteration in ubiquitin proteasomal function. [22,23,26] AMD could be one of the first manifestations of the development of dementia. The loss of visual inputs leads to reduced activation of central sensory pathways, resulting thus in an increased risk of cognitive damage to brain structures and the development of dementia. [27] The early diagnosis of cognitive brain degeneration and Alzheimer’s disease can be speculated using bloodless optical coherence tomography (OCT) in the retina. Parameters such as the reduced thickness of the retinal fiber nerve layer (RFNL) and the retinal ganglion cell layer, assessed with OCT, are pronounced in AD patients. [28] Other significant causes of visual impairment, such as cataracts, diabetic retinopathy (DR), and glaucoma, have been investigated in regard to cognitive decline and dementia, but with controversial results. [29-31] With this study, we aimed to examine if all the aforementioned SNP’s could also become a biomarker for AD. Another aim was to supplement the overall genotyping record of the ARMS2 and CFH polymorphisms that predecessor Greek studies conducted. [22,23] Despite our accurate AD sample, respecting the latest criteria for the diagnosis assessment, our results reach no statistically significant correlation for CFH [Odds Ratio=0.90, 95%C.I.=1.54-1.49, p=0.68] or the ARMS2 gene [OR=1.29, 0.60-2.77, p=0.5].
Nevertheless, our results are consistent with some reports where polymorphisms implicated in AMD are not associated with AD and, therefore, AMD has not been considered a precursor disease in dementia development. [32-34] A possible explanation might be that neuronal pathways remain unclear regardless of the common pathobiological mechanisms between AMD and AD. Similar alterations in the homeostasis of both diseases may lead to cerebral degradation but not necessarily to senile degeneration and/or senile dementia. An alternative explanation might be the reversed interpretation of the apolipoprotein E encoding between AD and AMD. The amyloid beta and APOE4 synergy in AD is well established. In AMD, in contrast to AD, APOE2 participates significantly in APOE overexpression and age onset of the disease, and it is considered a risk factor compared to APOE4’s protective role. [35] This hypothesis highlights the theory that APOE interacts differently with lipoprotein metabolism and transport. Also, in AMD, APOE is an abundant element that enacts in subretinal macrophage accumulation and retinal inflammation response and, thus, in drusen evolution. [36,37] Furthermore, APOE inhibits component pathways in the retina environment after interacting with a membrane pore. This membrane pore is formatted by an assembled effector of the complement pathway known as the membrane attack complex (MAC), and consequently, APOE is considered an important modulator of the complement cascade. [38,39] In addition, according to Scholl et al., ARMS2 and CFH polymorphisms can be considered risk elements to the predisposition of geographic atrophy but may not be to the progression of the disease once late AMD is already evolved [40].
The common genetic factors might seem insufficient for an association between the two neurodegenerative diseases. Moreover, a bigger sample in our study that cross-examines prevalent biomarkers for both diseases might have conducted a more concrete hypothesis and, thus, a better understanding of their pathogenesis. As mentioned, AD and AMD are both heterogeneous disorders; therefore, their complexities may be an ideal model for gene detection with novel next-generation sequencing (NGS) methods. It might be resourceful to examine the association of clinical variants of AD, such as logopenic aphasia in AMD, and other mutations or polymorphisms in genes involved in the common mechanisms of both diseases. Also, regulating Complement Factor H (CFH) in Alzheimer’s disease (AD) may redirect our therapeutic perspectives to novel treatment strategies that have not yet been considered. The link could be CFH alterations in expression, not CFH polymorphisms per se. MicroRNAs, such as miR-146a, could have more importance since they have been shown, in many studies, to lower CFH expression. [41] Other studies also presented that several miRNAs loci (miR-101, miR-20a, miR-17, miR-9-1, miR-7, miR-342, miR-410, miR-126) regulate the process of Aβ accumulation in drusen and the brain. Similarly, they interplay with the retina’s vascularisation and AMD’s pathogenesis through the apoptosis pathway. In the same vein, miRNA upregulation, synergistically with cytokines and beta-amyloid peptides, is involved, e.g., in downregulating the CFH expression.
Reduced CFH expression leads to an inflammatory response and, therefore, to drusen formation. This combinatorial effect targeting the disease’s pathogenesis may be used as a biomarker to diagnose and a possible therapeutic tool in both AMD and AD. [12,42] In conclusion, our results report no association of ARMS2 and CFH SNP with AD, even under adjustments for gender. AMD and AD share similarities, but their underlying genetic mechanisms may differ in the retina compared to the brain. However, this pilot study should be further extended to sub-groups of the two diseases and other genes involved in common mechanisms of pathogenesis in both disorders.
We thank all the participants and their families for contributing to this study.
The authors declare no conflict of interest.


