Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Sayed Tanveer Abbas Gilani1, Dilshad Ahmed Khan2*, Muhammad Amjad Pervez2, Amer Rauf1, Zujaja Hina Haroon2, Mohsin Saif1 and Naseer Ahmad Samore1
Received:March 09, 2023; Published:March 20, 2023
*Corresponding author: Dilshad Ahmed Khan, Prof of Pathology, National University of Medical Sciences, The mall, Rawalpindi, Pakistan
DOI: 10.26717/BJSTR.2023.49.007787
Background: Diagnosis of the coronary artery disease (CAD) in patients with angina is a challenge in
clinical settings without invasive coronary angiography. Our objective was to find out non-invasive molecular
diagnosis by relative expression of micro-RNA (miRNA) panel for CAD in the patients with angina.
Methods: Diagnostic accuracy study was conducted in tertiary care hospitals of Rawalpindi, Pakistan. A total
of 113 subjects, consisting of 58 cases, and 55 controls presented with chest pain, negative for Troponins,
and were having coronary artery stenosis >50%, and <50% on angiography, respectively. miRNA-21, miRNA-
33a, miRNA-133a, miRNA-145, and miRNA-146a were analyzed in blood with applied biological material kit
protocol of Canada by polymerase chain reaction on Rotor-Gene.
Results: In patients of CAD, relative expression of miRNA-33a, miRNA-133a, and miRNA-146a were
significantly (P<0.05) up-regulated, while miRNA-21, and miRNA-145 were non-significant in comparison
to controls. Receiver operating characteristic (ROC) curve analysis showed significantly (P<0.05) high area
under curve (AUC), (95% CI) along with sensitivity and specificity of miRNA-33a 0.89(0.83-0.94), 86% &
78%; miRNA-133a 0.68(0.59-0.78), 60% & 53%, and miRNA-146a 0.79(0.70-0.87), 74% & 62%, respectively.
CombiROC analysis of miRNA-33a, miRNA-133a, and miRNA-146a correctly identified CAD patients with
angina (AUC=0.93, sensitivity 82%, specificity 90%). Significantly (P<0.05) positive correlation was seen for
miRNA-33a (r=0.59), miRNA-133a (r=0.36), and miRNA-146a (r=0.37) with severity of CAD.
Conclusion: The panel of miRNA (miRNA-33a, miRNA-133a, and miRNA-146a) can be utilized as noninvasive
diagnostic biochemical marker for differentiating CAD patients from healthy individuals.
Keywords: Angina; Coronary Artery Disease; Diagnostic Accuracy; Micro RNA
Abbreviations: CAD: Coronary Artery Disease; miRNA: Micro-RNA; NIHD: National Institute of Heart Diseases; AFIC: Armed Forces Institute of Cardiology, NIHD: National Institute of Heart Diseases, NUMS: National University of Medical Sciences; AFIP: Armed Forces Institute of Pathology; HTN: Hypertension, DM: Diabetes Mellitus; VSMC: Vascular Smooth Muscle Cell; ROC: Receiver Operating Characteristic
The most common condition and cause of death in the developed world is coronary artery disease (CAD) [1]. Around the world, prevalence of CAD is 10 % [2], and in Pakistan, it is approximately 17 % [3,4]. The pathogenesis of CAD is complicated and includes endothelial cells (ECs) dysfunction, cholesterol uptake, chronic inflammatory reactions, and vascular smooth muscle cell (VSMC) proliferation. Emerging molecular technologies and multiple experimental studies on atherosclerosis are rapidly transforming the approaches that can be utilized to diagnose CAD [5]. Clinically CAD can present as angina. The discomfort or pain known as angina is brought on when the cardiac muscles do not receive enough oxygenated blood. It might feel like pressure or tightening in the ribcage. There may be discomfort in the back, neck, jaw, shoulders, or arms. Additionally, it might cause shortness of breath or even indigestion [6,7]. According to the recommendations of the American College of Cardiology (ACC)/American Heart Association (AHA), angina is classified as either stable angina (SA) or unstable angina (UA) [8,9]. The gold standard for confirming CAD in the patients with angina is invasive coronary angiography. Other diagnostic options such as stress tests have limited values in differentiating angina. Micro Ribo-Nucleic Acid (miRNA) can be used as non-invasive biochemical marker for diagnosis of the CAD in patients with angina. For patients living in remote areas, it is not possible to do angiographies for all patients of angina. But whole blood can be stored in preservatives and transported on ice to the molecular labs for analysis of miRNA. In order to prevent myocardial infarction (MI) and other cardiac complications, CAD must be diagnosed in angina patients as soon as possible.
Micro RNA is single-stranded noncoding RNA molecule of 18 to 22 nucleotides present in the cell cytoplasm [10]. By binding to the 3’UTR (untranslated region) of messenger ribonucleic acid (mRNA), miRNA can control gene expression at the post-transcriptional level. This can either prevent protein synthesis from mRNA or promote mRNA degradation [11]. It has been noticed that miRNA controls nearly 60% of human protein-coding genes, and each miRNA targets numerous mRNA [12]. The majority of genes, which are involved in the pathology of CAD, have putative binding sites for miRNAs, which are suspected of playing a role in this pathology [12]. Major scientific advances have concentrated on identifying miRNA as a diagnostic, prognostic, and therapeutic biochemical marker of CAD in the quest to find out non-invasive diagnosis of CAD [12,13]. Although many biological molecules, such as peptides, proteins, cytokines, and different metabolites, are being investigated as biochemical indicators for CAD, but their sensitivity and specificity are less than that of miRNAs [14,15]. Circulating miRNA may be the best biomarker for cardiovascular diseases because they are present in plasma and other body fluids as stable molecules that are protected from endogenous ribonuclease (RNAse) activity [12].
Out of various miRNAs studied to be involved in the developement of CAD, miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA-146a can be selected to cover the pathogenesis of atherosclerosis. miRNA-21 expression increases in ECs dysfunction, VSMC proliferation, oxidative stress, inflammation-based atherosclerosis, and insulin insensitivity [16]. miRNA-33a expression up-regulates in atherosclerosis and progression by lipid metabolism, glucose homeostasis and in the inflammatory pathways involving CAD [17,18]. In CAD, increased miRNA-133a is released from the ischemic myocardium into the circulation. miRNA-133a up-regulates in VSMC proliferation as well [19]. The abnormal proliferation of VSMCs is suppressed by miRNA-145, hence decrease in miRNA-145 can be a marker in prediction, progression, and severity assessment of CAD [20]. miRNA-146a is expressed in vascular endothelial cells, monocytes/macrophages, and smooth muscle cells, and it controls the inflammatory reaction that leads to atherosclerosis [21]. miRNA-146a over-expresses in the atherosclerotic diseases due to inflammation and VSMC proliferation [22]. Cardiac panel of miRNAs involved in complete pathogenesis of atherosclerosis can be used for accurate diagnosis of the CAD in patients with angina.
Currently, the clinical history is used to assess CAD in angina patients and angiography being invasive, expensive, difficult procedure, and not easily available in secondary and some tertiary care hospitals, is the only way to confirm the diagnosis of CAD. Up till now, no non-invasive biochemical marker exists in practice for the diagnosis of CAD in the patients with angina. The researchers have done several studies by using different miRNAs for diagnosis of CAD and dysregulation of few miRNAs are found in different studies that reported controversial results [12]. However, the panel of miRNAs involved in the pathogenesis that covers all the aspects of atherosclerosis can be used for accurate diagnosis of CAD is yet to be validated. The aim of this study was to; determine miRNA expression profiles of miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA-146a for early diagnosis of atherosclerosis in patients with angina; find out diagnostic accuracy of miRNA panel (miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA-146a) in comparison to angiography for CAD in the patients with angina; determine correlation of miRNA with severity of stenosis and extent of CAD by Gensini Score (GS).
This diagnostic accuracy study was carried out at Armed Forces Institute of Pathology (AFIP), National University of Medical Sciences (NUMS) in conjunction with Armed Forces Institute of Cardiology (AFIC) & National Institute of Heart Diseases (NIHD), NUMS, Rawalpindi, Pakistan, from October 2021 to September 2022 after obtaining approval of institutional ethical review board (IERB), AFIC IERB # 9/1/R&D/2020/50. A total of 113 individuals with a history of angina (based on ACC / AHA guidelines), presenting to AFIC, Rawalpindi, were recruited in the study [19]. Among these, 58 CAD patients of both genders, 25 to 70 years of age, with a history of any duration of angina, negative for Troponin-I (Trop-I) and had > 50% stenosis in any of the coronary artery on coronary angiography were selected by consecutive sampling [7,23], and 55 controls of almost similar age groups with no history of CAD and on angiography, coronary artery stenosis less than 50% were included [24,25]. Informed consent was obtained from all included individuals. Subjects with a documented history of Prinz-metal angina, MI, congestive heart failure, atrial fibrillation, history of either percutaneous intervention or coronary artery bypass grafting, with known history of renal or hepatic dysfunction, suffering from acute infection and chronic inflammatory disorders from past 6 weeks, were excluded from the study. In the research proforma, information about age, gender, blood pressure, body mass index (BMI), history of cardiovascular risk factors like smoking, dyslipidemias, hypertension (HTN), diabetes mellitus (DM), family history of CAD, and drug history was noted.
Coronary Angiography
Coronary angiography was performed on all of the included individuals to confirm the existence of CAD. According to ACC/AHA recommendations, 2 cardiologists separately assessed each patient’s angiography [7,19]. The severity coefficient, which gave scores of 1, 2, 4, 8, 16, and 32 to reductions of 25%, 50%, 75%, 90%, and 99% to each coronary stenosis based on the degree of luminal narrowing, was multiplied by coefficient determined by the myocardial area supplied by that segment, to determine the GS. Each integral lesion was scored by the degree of stenosis multiplied by the score of lesions location, and finally each patient’s GS was the sum of all integral lesions scores [24,26].
Laboratory Tests
Whole blood was collected from all patients of CAD and controls, in ethylene diamine tetra acetic acid (EDTA) and gel tubes to be tested for high sensitivity Troponin-I (hs-Trop-I), serum total cholesterol, triglycerides, low-density lipoprotein cholesterol, highdensity lipoprotein cholesterol, plasma glucose fasting, blood counts, creatinine and alanine transaminase {on automated chemistry analyzer (Architect Ci 4100 System Abbott, USA)} and micro RNA panel (miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA- 146a) on Rotor-Gene.
RNA Extraction
Whole blood in EDTA sample was mixed in 1 x RNA shield to preserve RNA and immediately stored at −80°C till analysis. RNA extraction was done by quick RNA whole blood kit of Zymo Research, USA (R-1201). To normalize the whole blood miRNA content, we supplemented caenorhabditis elegans miR-39-3p spike in control to all samples during the extraction. After extraction, optical density (OD) was measured by nano drop spectrophotometer; ratio of 260 nm / 280 nm more than 1.8 showed that RNA was pure. All RNAextracted samples were stored in the aliquots at − 80°C till analysis.
Reverse Transcription
RNA template (200 ng/reaction) from each sample was used for complementary deoxyribo nucleic acid (cDNA) synthesis by using kit protocol G899 of applied biological material (ABM), Canada. All cDNA samples were stored in the aliquots at − 80°C till analysis.
Quantitative Polymerase Chain Reaction (qPCR)
Bright Green miRNA qPCR MasterMix-ms of ABM-Canada was used in real-time PCR (Rotor-Gene of Qiagen, Germany) according to the kit protocol. A total of 20 μl of each reaction well was containing 0.6 μl of universal reverse transcribed miRNA, 0.6 μl each of forward assay primer (miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA-146a) or control miR-39-3p primer, 2.5 ul (1:10 dilution) of template cDNA, 6.3 μl of molecular-grade water and 10 μl of the master mix per well. Thermal protocol was containing 10 minutes (min) of enzyme activation at 95°C, then 40 cycles of the denaturation at 95°C for 10 seconds (sec), annealing at 63°C for 15 sec and the extension at 72°C for 5 sec followed by detection. Real-time PCR was performed in duplicate. miRNA was considered present / expressed when CT (cyclic threshold) values were less than 35.
Relative Expression
miRNA relative expression was determined using 2−ΔCT calculation method. Relative expression of circulatory miRNA after normalization to spike in control cel-miR-39-3p was calculated using comparative CT method as given below [27].
Target Sequences of miRNAs
Target sequences of miRNA-21, miRNA-33a, miRNA-133a, miRNA-145, and miRNA-146a were shown in (Table 1).
Statistical Analysis
Using AUC 0.779, Alpha (type 1) errors 0.05, Beta (type 2) errors 0.10, and 50% null hypothesis, sample size was computed by MedCalc. The result was 42 samples, each for the cases and controls [22]. The statistical analysis was performed using SPSS (Statistical Package for Social Sciences) version 22. Kolmogorov-Smirnov (K-S) test was used to identify the type of data. The qualitative variables were converted into frequencies and percentages. The chi-square test was used to analyze categorical variables. The quantitative data was given as means and standard deviations (SD) for the parametric data, and median and interquartile ranges (IQR) for the non-parametric data. The independent sample t-test was used to compare continuous variables that were parametric, and the Mann Whitney U test was used to analyze non-parametric data. To assess the diagnostic accuracy, a Receiver Operating Characteristic (ROC) curve was drawn for miRNA relative expression. CombiROC analysis was carried out online on combiroc.eu by combining ROC analysis of miRNAs having better diagnostic accuracy. The relationship between relative expression of miRNA and the severity of CAD as determined by coronary artery stenosis on angiography and GS was measured by Spearman’s correlation. Significant results were defined as having two-tailed P value less than 0.05.
Out of 113 subjects, 58 were patients of CAD, and 55 were controls. The age of the patients with CAD was 55±12 years and of controls was 49±12 years. According to Asian population criteria, the vast bulk of study participants had an overweight condition with a BMI of more than 23 kg/m2. The types of drugs used by the study participants were also noted. They were following the doctor’s recommendations for a typical diet and routine exercise (Table 2). In patients of CAD, median (inter quartile range / IQR) of coronary artery stenosis 69 (59 – 90) % was significantly (P < 0.001) more than controls 15 (0- 25) %. In patients with CAD, median (IQR) of GS, 17 (11-38) was significantly (P < 0.001) more than controls 1.5 (0-3.5). The patients with angina had significant (P < 0.001) up-regulation of relative expression {median (IQR)} as compared (vs) to controls for miRNA- 33a {15.98 (2.66-35.17) vs 0.97 (0.29-2.08)}, miRNA-133a {22.8 (9.1- 82.7) vs 13.4 (5.5-20.3)} and miRNA-146a {2.03 (0.28-8.07) vs 0.09 (0.03-1.67)}. While non-significant up-regulation of miRNA-21 {0.07 (0.02-0.57) vs 0.06 (0.001-0.40)} and down-regulation of miRNA-145 {0.65 (0.17-1.34) vs 0.74 (0.29-1.95)} were observed in CAD patients with angina as compared to controls, respectively (Figure 1).
Note: “BMI: Body mass index; Total-C: Total cholesterol; HDL-c: High-density lipoprotein-cholesterol; LDL-c: Low-density lipoprotein-cholesterol; VLDL-c: Very low-density lipoprotein-cholesterol; hs-Trop-I: high sensitivity Troponin-I; PGF: Plasma glucose fasting; ALT: Alanine transaminase; HTN: Hypertension; DM: Diabetes mellitus; S/C: Sub-cutaneous; ACE: Angiotensin converting enzyme. Data were expressed as number (percentage) = n (%), mean ± SD and median (interquartile range); Chi-square test for qualitative, independent sample t-test for quantitative parameters and nonparametric data by Mann-Whitney U test* analysis between patients of CAD and control groups; P value < 0.05 was significant”.
ROC curve analysis showed good diagnostic potential for miRNA- 33a, miRNA-133a, and miRNA-146a. CombiROC analysis showed that combination of miRNA-33a, miRNA-133a, and miRNA-146a {AUC = 0.93, sensitivity (82%), specificity (90%), positive likelihood ratio (LR) 8.2, negative LR 0.2} correctly identified the CAD patients with angina in comparison to controls (Table 3). This study found significantly (P < 0.05) positive correlation of relative expression of miRNA-33a (r = 0.59, r = 0.54), miRNA-133a (r = 0.36, r = 0.34), and miRNA-146a (r = 0.37, r = 0.38), with coronary artery stenosis and the GS, respectively (Table 4). miRNA-33a relative expression of 2.12 to 10.42 had categorized the patients into moderate CAD (50 to 70 % stenosis) and for severe CAD (more than 70 % stenosis) relative expression was more than 10.42. miRNA-133a relative expression 14.3 to 23.5 in moderate CAD and for severe CAD more than 23.5. miRNA-146a relative expression 0.30 to 1.05 in moderate CAD and more than 1.05 in severe CAD.
Table 3. Diagnostic accuracy analysis of relative expression of miRNAs in patients of CAD (n=58) as compared to controls (n=55).
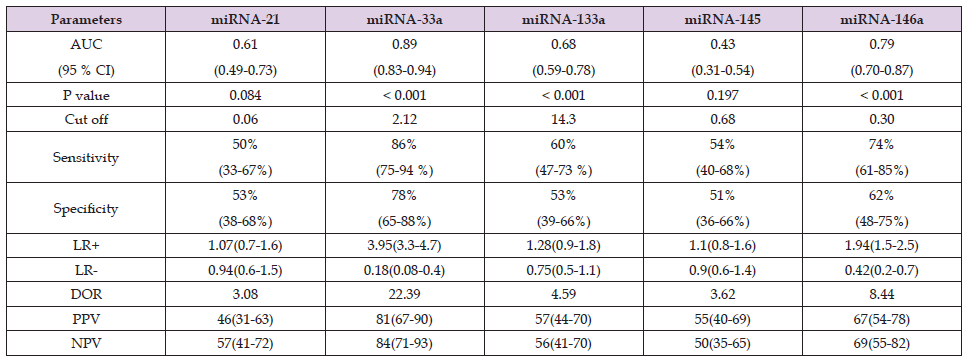
Note: “miRNA: Micro RNA; CAD: Coronary artery disease; AUC: Area under curve; CI; Confidence interval; LR: Likelihood ratio; DOR: Diagnostic odds ratio; PPV: Positive predictive value; NPV: Negative predictive value; P value < 0.05 was significant”.
Table 4. Spearman’s correlations of relative expression of miRNAs with stenosis of coronary artery and Gensini Score (N=113).
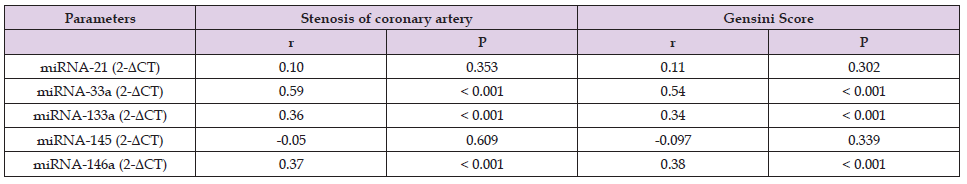
Note: “miRNA: Micro RNA; CT: Cyclic threshold; r: Spearman’s correlation; P value < 0.05 was significant”.
This novel study emphasizes the use of miRNA panel for the diagnosis of CAD in patients with angina. In literature, individual miRNAs were studied for CAD, especially for MI but lesser studies were available for early stages of CAD (stable or unstable angina), by different authors with variable results. This is the first study in which use of micro-RNA panel involved in complete pathogenesis of the atherosclerosis to diagnose patients with angina was established in comparison to the angiography. miRNA is helpful to diagnose the CAD in patients with angina and severity assessment of atherosclerosis. In absence of the myocardial necrosis (increased plasma levels of Troponins), there is presently no recognized biochemical marker in practice that can be used as confirmatory diagnosis of the CAD in patients with angina. Inflammatory biomarkers of the atherosclerosis are not cardiac specific; hence their diagnostic accuracy is low for the diagnosis of CAD. Notably, it has been shown that failure to diagnose cardiac ischemia increases both short and mid-term mortality [9]. Therefore, it is a compelling need to identify novel noninvasive cardiac biomarkers that are not dependent on myocardial necrosis. In circulation, miRNAs exist in extremely stable forms. These miRNAs are packed in the transport particles, such as micro-particles, exosomes, and protein complexes, which can protect miRNA from degradation [12]. Hence miRNA can be ideal non-invasive biochemical marker in diagnosing patients of CAD.
The present study had shown diagnostic potential of miRNA panel for CAD and highly significant positive correlation of relative expression of miRNA-33a, miRNA-133a, and miRNA-146a with coronary artery stenosis and GS. The current study demonstrated significant diagnostic value of miRNA-33a for CAD patients with angina, better than published study that was revealing AUC 0.80 (cutoff = 9.91), sensitivity (72%), and specificity (67%), miRNA-33 upregulated in CAD patients was also found positively correlated with severity of atherosclerosis [17]. miRNA-33a was found up-regulated in cases of CAD having atherosclerosis due to dyslipidemias, inflammation and DM [18]. By binding to 3’UTR of the genes that are involved in the reverse transport of cholesterol, such as ATP-binding cassette transporters, miRNA-33a increases the cholesterol content in tissues by repressing the expression of these genes. According to studies, ATP binding cassette transporter (ABCA1) mediates the delivery of cholesterol from peripheral tissues to apolipoprotein A-1 (ApoA-1) and is crucial in the reverse cholesterol transport pathway, where cholesterol is given from peripheral tissues to liver, then it may either be transformed into bile acids or excreted in bile [17]. The macrophages’ ability to accumulate cholesterol is inhibited by ABCA1. By increasing the miRNA-33a expression, level of ABCA1 is decreased, hence causing reduction in cellular cholesterol efflux to the Apo A-1. Up-regulation of miRNA-33a is also involved in tumor necrosis factor (TNF) receptor stimulatory pathway of inflammation leading to atherosclerosis [17]. miRNA-33a is involved in early development of atherosclerosis, hence more specific in early diagnosis of angina, as demonstrated in current study.
In our study, significant elevation of miRNA-133a was found for CAD patients with angina as compared to controls, but with lesser diagnostic accuracy than available studies in literature. miRNA-133a was found to be a marker of coronary artery stenosis with cut off = 4.56, AUC = 0.90, sensitivity (86 %), and specificity (92 %) in acute coronary syndrome (ACS) patients [19]. AUC value 0.91 was also observed in UA for miRNA-133a [9]. miRNA-133a may be used as non-invasive biomarker for CAD due to its role in the diagnosis and severity assessment [19,27]. miRNA-133a is cardiac-myocyte specific miRNA that is up-regulated in ischemia, having better-discriminating potential for CAD than hs-Trop-I because miRNA-133a is also upregulated in VSMC proliferation and ideal biomarker covering both cardiac-myocyte ischemia and atherosclerosis for diagnosis and severity assessment of CAD. As ischemia and VSMC proliferation was found in severe stages of atherosclerosis that is why miRNA-133a had discriminating values for severe cases of angina. In this study, significant elevation of miRNA-146a was observed for CAD patients with angina as compared to controls, as similar to published study in which AUC of miRNA-146a was found 0.78 (P < 0.001) for the diagnosis of CAD [22]. GS for the assessment of CAD severity found that miRNA-146a was positively correlated with GS, indicating that miRNA-146a was elevated according to severity of the CAD [22,28,29].
miRNA-146a has better diagnostic potential for CAD than existing biomarkers of inflammation because miRNA-146a is up-regulated in inflammation and VSMC proliferation involved in atherosclerosis. Cytokines like interleukin-1 and TNF induce miRNA-146a expression in a delayed kinetic way. By specifically targeting the upstream adaptor proteins TNF receptor-associated factor 6 and interleukin 1 receptorassociated kinase 1 or 2 (IRAK1/2), miRNA-146a also inhibits the production of EC adhesion molecules. Multiple genes in the toll-like receptor 4 (TLR4)/nuclear factor kappa B (NF-kB) pathway, a crucial immune and inflammatory mechanism that controls inflammatory response, are targeted by miRNA-146a [22,28]. Inflammation presents in early stages of atherosclerosis while VSMC proliferation occurs in severe stages of atherosclerosis that’s why miRNA-146a can be ideal non-invasive biomarker for discrimination of healthy controls from patients with angina in early as well as late stages of CAD.
Contrary to our study; in few published studies, AUC 0.86 (P = 0.0001) for ACS and 0.78 (P = 0.0001) for SA from healthy subjects were the results of the diagnostic accuracy of miRNA-21 by ROC curve [30]. Positive correlation in expression of miRNA-21 with severity of CAD was found, because of its role in inflammation, oxidative stress, and insulin insensitivity [30]. Reduced expression of miRNA-145 had relatively good diagnostic accuracy, according to ROC curve created using the expression levels of this miRNA, with an AUC of 0.85, sensitivity (84%), and specificity (83%) at a cut-off = 5.6 [31]. miRNA-145 AUC value of 0.78 was also observed in UA as compared to controls [9]. The decrease in miRNA-145 can be useful marker of severity assessment of CAD because it inhibits VSMC proliferation [20,32]. Our previous study showed that in the patients of CAD, inflammatory biomarkers like TNF-alpha and NF-kB had betterdiscriminating potentials than hs-CRP and IL-6, but these biomarkers can be elevated in other inflammatory conditions as-well [33]. While in cardiac-specific micro-RNAs, miRNA-33a has better-discriminating potential for CAD in patients with angina, than miRNA-133a, miRNA- 146a, and other inflammatory biomarkers of atherosclerosis. miRNA- 33a and miRNA-146a have better diagnostic odds ratio, positive LR, negative LR, positive predictive values, and negative predictive values than other miRNAs under study. Relative expression of miRNA-33a, miRNA-133a, and miRNA-146a categorized the patients into minor, moderate, and severe CAD.
CombiROC analysis showed that combination of miRNA-33a, miRNA-133a, and miRNA-146a correctly identified the CAD patients with angina in comparison to controls. As in atherosclerosis of coronary arteries, miRNA-33a is up-regulated due to dyslipidemias, inflammation, and DM; miRNA-133a is up-regulated due to cardiacmyocytes ischemia and VSMC proliferation; miRNA-146a is upregulated in inflammation and VSMC proliferation; so the panel of these three micro RNAs (miRNA-33a, miRNA-133a, and miRNA- 146a) covering all aspects of the pathogenesis of atherosclerosis have excellent diagnostic potential for CAD in patients with angina.
Limitations
Whole micro-RNA panels were not studied; only few selected cardiac-specific miRNAs were analyzed. Next-generation sequencing may be used for whole genome sequencing. Messenger RNA involved in pathogenesis of the atherosclerosis may be analyzed in future studies. In this diagnostic accuracy study, only internal validation of results was done with angiography. External clinical validation for prediction of the CAD in patients with angina is recommended to be carried out in further studies.
The panel of miRNA (miRNA-33a, miRNA-133a, and miRNA-146a) has diagnostic value in the early detection of CAD in angina patients. This panel of miRNA may be used as non-invasive biochemical marker prior to coronary angiography in order to distinguish healthy individuals from CAD patients. In medical settings without access to cath lab equipment, these miRNAs can also be used for the diagnosis of CAD.
Our study was supported by National University of Medical Sciences, Rawalpindi.
None of the authors revealed any conflicts of interest.


