Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Akinaw Wagari*
Received:January 02, 2023; Published:February 09, 2023
*Corresponding author: Akinaw Wagari, College of Veterinary and Agriculture, Addis Ababa University, P. O. Box 34, Bishoftu, Ethiopia
DOI: 10.26717/BJSTR.2023.48.007665
Livestock carry large numbers of protozoa in their stomachs and intestines, the vast majority of which are entirely harmless. Cryptosporidium infects various livestock, resulting in significant economic losses. Some species of protozoa, however, are significant as causes of disease in domestic cattle, sheep and poultry, or because of their potential for zoonotic transmission. The hosts ranges and pathogenicity is species variable. Sheep are infected with three main Cryptosporidium species; Cryptosporidium parvum, Cryptosporidium ubiquitum and Cryptosporidium xiaoi and humans are mostly infected with Cryptosporidium parvum (the most zoonotic species) and Cryptosporidium hominis where Cryptosporidium hominis is commonly associated with human infection while Cryptosporidium parvum is linked with infection in animals, especially young ruminants. According to a recent study, more than 44 Cryptosporidium species have been identified and more than 70 genotypes that are recognized as valid on the basis of morphological, biological and molecular data. The infection is transmitted to animals and humans orally through the ingestion of sporulated oocysts. In particular, neonatal calves and lambs are vulnerable to Cryptosporidium infection and shed millions of oocysts, resulting in enormous environmental contamination and a risk of infection to other animals and humans. And also, untreated manure use as fertilizer for vegetable cultivation also poses a public health threat. Since parasitological techniques are poor, the identification of species and subtypes of Cryptosporidium is dependent on molecular techniques. Both humans and animals had a high risk of infection from contaminated feed and water due to improper handling of manure on dairy farms and rural farmers’ households, so dairy farmers and farmer households should handle, treat and store animal manure properly in order to reduce the risk of infection to both animals and humans.
Keywords: Cattle; Cryptosporidiu; Human; Lamb; Zoonotic
Gastrointestinal parasites are considered as one of the most significant constraints in livestock sector. Damages inflicted to health and productivity includes loss in body weight, poor reproductive performance, digestive disturbances and emaciation for long period. It has been established that parasitic infections result in considerable economic losses in livestock sector [1]. Livestock carry large numbers of protozoa in their stomachs and intestines, the vast majority of which are entirely harmless. Considering its epidemiological profile, Cryptosporidium infects various livestock, resulting in significant economic losses. Some species of protozoa, however, are significant as causes of disease in domestic cattle, sheep and poultry, or because of their potential for zoonotic transmission [2]. Cryptosporidiosis is a major diarrheal disease caused by a protozoan parasite of the genus Cryptosporidium, family of Cryptosporididae, Order Eucoccidiorida, class of Coccidia and phylum Apicomplexa. The parasite infects epithelial cells in the microvillus of gastrointestinal tract of all classes of vertebrates and causes severe chronic and even fatal diarrhea with mal-absorption and dehydration [3]. These organisms are one of the most prevalent parasites that infect domesticated cattle and sheep. The genus Cryptosporidium consists of eukaryotic protozoal intracellular parasites and is classified as a member of the phylum Apicomplexa. The four major Cryptosporidium species that infect cattle are Cryptosporidium parvum, Cryptosporidium bovis, Cryptosporidium ryanae and Cryptosporidium andersoni [4].
According to a recent study, more than 44 Cryptosporidium species have been identified and more than 70 genotypes that are recognized as valid on the basis of morphological, biological and molecular data [5]. The hosts ranges and pathogenicity is species variable. Sheep are infected with three main Cryptosporidium species; Cryptosporidium parvum, Cryptosporidium ubiquitum and Cryptosporidium xiaoi and humans are mostly infected with Cryptosporidium parvum (the most zoonotic species) and Cryptosporidium hominis where Cryptosporidium hominis is commonly associated with human infection while Cryptosporidium parvum is linked with infection in animals, especially young ruminants [6]. Among the species, Cryptosporidium parvum is the most common species of medical and veterinary importance. Most people and animals infected with Cryptosporidium parvum develop immunity and recover from that infection. However, the disease is persistent and life threatening if there is immunologic impairment [7]. The infection is transmitted to animals and humans orally through the ingestion of sporulated oocysts [8]. The sexual and asexual life cycles are completed in the same host (monoxenoes) and have a unique location within the host cell, situated between the cytoplasm and the cell membrane [9].
Calves are primarily infected via the fecal-oral route and it takes less than 50 oocysts to infect a healthy calf [10]. Infection can rapidly spread from calf to calf when animals are communally housed and overcrowded or from cow to calf via the udders when they are contaminated with infected calf feces in the lying area of the dams [11]. These oocysts are resistant to the environment and remain infective for months in cold water or dump, cool environment [12]. In particular, neonatal calves and lambs are vulnerable to Cryptosporidium infection and shed millions of oocysts, resulting in enormous environmental contamination and a risk of infection to other animals and humans. Meanwhile, asymptomatic weaned and adult cattle also excrete oocysts into the environment. According to [13] a single adult bovine might possibly excrete more than 36 million oocysts every day, but also the sub-clinically infected ewes are also a source of infection for lambs, especially during the peri-parturient period. So, without adequate control, this contamination represents a human health hazard because infected animals could shed up to 107 oocysts per gram of feces [14].
Oocysts are resistant to environmental conditions and survive for a months in environments and animal manures under cool and wet conditions. Infected animal manure also serves as a significant reservoir for Cryptosporidium infection [15]. Surface transfer from land-applied manures or leaching through the soil to groundwater is two additional mechanisms of transfer of the pathogen to drinking or recreational water, in addition to direct fecal deposition. Runoff from polluted field might act as a vehicle for Cryptosporidium oocysts to enter water sources. As a result, cattle farms might be a major source of Cryptosporidium infection for humans and other animals [7]. Untreated manure use as fertilizer for vegetable cultivation also poses a public health threat. Additionally, intense rainfall results in surface water runoff, which can transport oocysts from farms into surrounding watersheds (water sources), resulting in high pathogen loads and more potential for human infection [16].
Clinical cryptosporidiosis is frequently not diagnosed, yet it has been incriminated as an important cause of diarrhea in neonates [17,18]. Clinically, the disease is characterized by anorexia and diarrhea, often intermittent, which may result in poor growth rate [19,20]. The severity of clinical disease may be associated with the animals’ immune and nutritional status [12]. It is also characterized by low morbidity which however may become severe when associated with other pathogens [21,22]. According to [11], although calves 1-3 weeks old seem to be most susceptible, cryptosporidium species has also been found in cattle over two years of age impairing rate of gain in feedlot cattle and milk production in dairy cattle [12]. A variety of methods is available for detection of Cryptosporidium species including microscopic, immunological and molecular techniques. Microscopic detection is based on finding the environmental and chemical resistant oocysts in fecal samples [22]. Oocysts may be demonstrated using Ziehl-Nielsen stained fecal smears in which the sporozoites appear as bright red granules [19].
The identification of species and subtypes of Cryptosporidium is dependent on molecular techniques. In many parts of Africa, the infrastructure for molecular characterization is not yet evolved and consequently studies on the distribution of Cryptosporidium species, genotypes and transmission routes are scanty in our country [23]. Various reports have indicated that the prevalence of cryptosporidiosis ranges from 6.25 to 39.65% in cattle in different parts of the world. But also, the prevalence of Cryptosporidium infection ranges from 10.8 to 27.8%, and 2.1 to 22.2%, in calves and lambs, respectively. On the other hand, [24] reported the highest prevalence (46%) of Cryptosporidium infection in humans having contact with animals. There are various factors that affect the prevalence of cryptosporidiosis including age, bedding type, hygiene, colostrum feeding, management practices, feed and water sources, diarrhea and climate [25]. In Ethiopia, where over 65 million cattle, 40 million sheep, [26] are raised under various agro-ecological zones and having over 120 million population, a few research work and review paper have been done on cryptosporidiosis in different parts of the country.
Objective’s
A. To review different parasitological and molecular techniques
that help us to rapidly diagnosis the disease in animals and
human.
B. To highlight the current status and zoonotic implication of
the disease in our country.
C. To show a research gap for the disease that will enable researchers
in the near future to show the importance of the disease in
our country in a broad manner.
Cryptosporidiosis in Calves
Only one species, Cryptosporidium parvum, causes disease in cattle and generally only in neonatal calves. The typical age for crypto is in the first 4 weeks of life. Clinical signs can range from mild scouring to calf death depending on the parasite burden, susceptibility and health status of the calves [27]. The higher the dose the more severe the symptoms, nutritional or energy stresses on the calf will make the symptoms more severe [28]. The life cycle of the parasite allows it to multiply rapidly in the host leading to the rapid spread of the disease as infected calves shed millions of parasites into the environment creating major breakdowns in calf rearing units and the parasite survives well in the environment and is not killed by most standard disinfectants. But also, the parasite survives best in wet or damp conditions and can survive for more than a year in damp environments having temperatures up to 600C and -200C. Steam cleaning is effective method of disinfection and currently there is no vaccine available and treatment options are limited [29]. The infective dose is 10 parasites! An infected calf can shed billions of parasites. One infected calf causes major contamination of the environment allowing infection to be spread to naive calves through contaminated bedding, feeding utensils or feed troughs [30]. Mixing ages is high risks, since older calves or any calf recovering from infection can infect younger calves and perpetuate the life cycle; which takes between 2 and 7 days to complete from the point of infection to shedding oocysts in the faces; this provide infection to be built up very quickly [31]. Treating affected calves is challenging. In severe cases the disease will cause death with typically a poor response to treatment in many cases [17]. So, prevention is the key by using medication like Halocur which can be used by mixing with milk or dosing by mouth for the first 7 days [32]. But, any treatment must be controlled with improved hygiene, frequent bedding and using correct disinfectants frequently enough to break the life cycle, because Crypto can leak out of the pen into the passageways and contaminate where you walk and this can be a common source of infection to other calves; so, pressure washing of hutches is essential to prevent cross contamination [30].
Parasitological Techniques in Calve:
Preparation of the Fecal Samples: A fecal sample is obtained from each animal directly from the rectum using a sterile plastic glove; then the sample is placed in a plastic cup and transported to the laboratory into an icebox to be examined within 2-3hr. The collected samples will be prepared and examined on the day of collection. The samples are transported and examined in Lab to determine the presence of Cryptosporidium infection [10].
Macroscopic Examination of the Fecal Samples: The fecal samples are examined macroscopically to detect abnormalities in consistency and color, the presence or absence of blood, the state of digestion and the presence of mucus or other unusual constituents according to the protocol described elsewhere [10].
Parasitological Examination of the Fecal Samples: The fecal samples are filtered through two layers of gauze to remove the coarse particles and stored in an equal amount of 2.5% potassium dichromate solution at 4◦C until the time of examination. All specimens are examined for Cryptosporidium oocysts under a microscope using a staining technique. The fecal samples are examined using the direct and saline smear methods according to the protocol described elsewhere [19].
Direct Smear Method: These steps are performed according to the protocol established elsewhere. Briefly, approximately 2mg of feces are mixed with a drop of physiological saline (0.85% NaCl) and placed on a clean slide. Then, the samples are thoroughly mixed until a uniform suspension was formed without fibers or gritty materials and covered with a 22 × 22 mm cover glass until the sample is evenly spread. The examinations are performed systematically and thoroughly using a 10× objective lens and confirmation will be made by switching to a magnification of 40× [33].
Simple Gravity Sedimentation Technique: Approximately 10g of feces are thoroughly mixed with 50ml of tap water in a 250ml beaker. Then, the suspension is strained through two layers of wet gauze into a sedimentation flask and the samples are allowed to sediment for 1hr. The supernatant is decanted carefully; the sediment is suspended by adding tap water and the material are left to sediment for 1hr. Then, the supernatant is decanted carefully again. Washing is repeated until a clear supernatant is obtained. A small portion of the sediment is placed on a glass slide using a long capillary pipette. The slide is covered with a cover slide and examined for parasites [22].
Flotation Method: The fecal samples (∼10g) are placed in a cup and mixed thoroughly with ∼50 mL of tap water using a spatula. The mixture is poured through a wire mesh screen with an aperture of 500μm to remove large lumps. The strained fluid is caught in a bowl. Then the suspension is transferred to a conical measure and filled with tap water to the top and allowed to settle for 30min. The supernatant was discarded carefully. The sediment is stirred and a 2ml sample is poured into a centrifuge tube. The tube is placed in a centrifuge. A saturated NaCl solution is added using a pipette until a convex meniscus stood above the top of the tube. A cover glass is placed on the tube, ensuring that no bubble is trapped under it. The tube is centrifuged at 2,000 rpm for 2min. The cover glass is removed and the sample is placed on a slide and examined under a microscope [20].
Examination of the Fecal Samples Using the mZN Method: Cryptosporidium is directly identified using the modified Kinyoun acid- fast stain (cold method). The films are fixed in absolute methanol for 2min and allowed to dry. The slides are flooded with Kinyoun’s Carbol-Fuchsin solution for 5min. The slides are rinsed briefly with 50% ethanol for 5s, rinsed thoroughly with water and decolorized with 1.5% sulfuric acid for 2min. Then, they are rinsed with water and drained; and counter-stained with methylene blue for 5min. The slides are rinsed again with water and allowed to air dry. The stained smears are systematically examined under a microscope at ×400 and ×1,000 magnification. Cryptosporidium species oocysts appear pink to red with spherical to ovoid bodies against a blue background [19].
Molecular Techniques in Calve:
Genomic DNA Extraction: This step involved the extraction of DNA from Cryptosporidium- positive fecal samples identified using microscopy and for example the QIAamp DNA MiniKit (Qiagen Co., USA) according to the manufacturer’s protocol. Following the extraction, DNA concentrations are measured using DNA/Protein Analyzer (Quawell Q 9000, USA) [34].
Nested PCR Procedure: this is performed as described by (Johnson et al., 1995). The primary PCR reaction contained 2μL of DNA in a 20μL reaction volume, and the secondary PCR re-amplified 5μL of the primary PCR product. Non-acetylated bovine serum albumin (Invitrogen, USA) at a final concentration of 400μg/ml is added to each PCR reaction. The primary and secondary protocols used are as follows: initial denaturation at 94◦C for 5 min; 40 cycles at 94◦C for 1 min, 55◦C for 1 min and 72◦C for 1 min; extension at 72◦C for 10 min. PCR amplifications are processed using for example BIO-RAD Thermal Cycler (BIO-RAD, Singapore). After amplification, the PCR products are visualized in a 1% agarose gel stained with RedSafe Gel electrophoresis (Intron) electrophoresis using Molecular Imager BIORAD, Singapore) [35].
Prevalence of Cryptosporidiosis in Calves in Different Parts of Ethiopia
Cryptosporidiosis in Lambs
In lambs infections appears to be age related with seasonal peaks of disease reported to coincide with birth peaks in spring and autumn. Although outbreaks in lambs are sporadic, mortality can be high. Cryptosporidium parvum is not host-specific and is in fact the second most common cause of diarrhoea in calves as well as in lambs [36]. Therefore it is conceivable an environment contaminated with oocysts during an outbreak in calves can cause infection in lambs using the same premises or grazing area [19]. Lambs as young as 3 days old can be affected which become depressed and reluctant to suck while the diarrhoea lasts for a week. Very young lambs soon become dehydrated and die, while in poor weather conditions lambs may die of hypothermia. The illness may last for up to 10 days, and relapses after apparent recovery are common [37]. In the face of an outbreak of cryptosporidiosis in lambs there is little that can be done other than to isolate affected stock, avoiding overcrowding or prolonged use of contaminated areas [38]. The tiny, fragile looking oocysts are infact remarkably tough and can survive in the environment for many months. Disinfection can be achieved by steam heat or by using Oocide (Antec International Ltd.), an ammonia-based oocysticide, which is particularly effective against Cryptosporidium oocysts [39]. As no specific ovine treatment measures are available at present, only the symptoms can be treated, by keeping lambs warm and giving rehydration therapy. However, in some cases, it is possible to use calf treatments, such as Halocur (Halofuginone lactate) [40].
Parasitological Techniques in Lamb
Preparation of the Fecal Samples: A fecal sample are obtained from each animal directly from the rectum using a sterile plastic glove; then the sample is placed in a plastic cup and transported to the laboratory into an icebox to be examined within 2-3hr. The collected samples will be prepared and examined on the day of collection. The samples are transported and examined in Lab to determine the presence of Cryptosporidium infection [10].
Macroscopic Examination of the Fecal Samples: The fecal samples are examined macroscopically to detect abnormalities in consistency and color, the presence or absence of blood, the state of digestion and the presence of mucus or other unusual constituents according to the protocol described elsewhere [10].
Parasitological Examination of the Fecal Samples: The fecal samples are filtered through two layers of gauze to remove the coarse particles and stored in an equal amount of 2.5% potassium dichromate solution at 4◦C until the time of examination. All specimens are examined for Cryptosporidium oocysts under a microscope using a staining technique. The fecal samples are examined using the direct and saline smear methods according to the protocol described elsewhere [19].
Direct Smear Method: These steps are performed according to the protocol established elsewhere. Briefly, approximately 2mg of feces are mixed with a drop of physiological saline (0.85% NaCl) and placed on a clean slide. Then, the samples are thoroughly mixed until a uniform suspension was formed without fibers or gritty materials and covered with a 22 × 22 mm cover glass until the sample is evenly spread. The examinations are performed systematically and thoroughly using a 10× objective lens and confirmation will be made by switching to a magnification of 40× [33].
Simple Gravity Sedimentation Technique: Approximately 10g of feces are thoroughly mixed with 50ml of tap water in a 250ml beaker. Then, the suspension is strained through two layers of wet gauze into a sedimentation flask and the samples are allowed to sediment for 1hr. The supernatant is decanted carefully; the sediment is suspended by adding tap water and the material are left to sediment for 1hr. Then, the supernatant is decanted carefully again. Washing is repeated until a clear supernatant is obtained. A small portion of the sediment is placed on a glass slide using a long capillary pipette. The slide is covered with a cover slide and examined for parasites [22].
Flotation Method: The fecal samples (∼10g) are placed in a cup and mixed thoroughly with ∼50 mL of tap water using a spatula. The mixture is poured through a wire mesh screen with an aperture of 500μm to remove large lumps. The strained fluid is caught in a bowl. Then the suspension is transferred to a conical measure and filled with tap water to the top and allowed to settle for 30min. The supernatant was discarded carefully. The sediment is stirred and a 2ml sample is poured into a centrifuge tube. The tube is placed in a centrifuge. A saturated NaCl solution is added using a pipette until a convex meniscus stood above the top of the tube. A cover glass is placed on the tube, ensuring that no bubble is trapped under it. The tube is centrifuged at 2,000 rpm for 2min. The cover glass is removed and the sample is placed on a slide and examined under a microscope [20].
Examination of the Fecal Samples Using the mZN Method: Cryptosporidium is directly identified using the modified Kinyoun acid- fast stain (cold method). The films are fixed in absolute methanol for 2min and allowed to dry. The slides are flooded with Kinyoun’s Carbol-Fuchsin solution for 5min. The slides are rinsed briefly with 50% ethanol for 5s, rinsed thoroughly with water and decolorized with 1.5% sulfuric acid for 2min. Then, they are rinsed with water and drained; and counter-stained with methylene blue for 5min. The slides are rinsed again with water and allowed to air dry. The stained smears are systematically examined under a microscope at ×400 and ×1,000 magnification. Cryptosporidium species oocysts appear pink to red with spherical to ovoid bodies against a blue background [19].
Molecular Techniques in Lamb
DNA Extraction and PCR Analysis: Potassium dichromate is washed off fecal samples with distilled water by centrifugation. Genomic DNA is extracted from 0.2ml of fecal slurry without further pathogen concentration using for example FastDNA SPIN Kit for Soil (BIO 101, MP Biomedicals, Carlsbad, CA, USA). DNA preparations are screened for Cryptosporidium spp. by using a small subunit (SSU) rRNA- based nested PCR, with DNA of C. baileyi as the positive control and reagent-grade water as the negative control. The detection limit of the approach will be ~10 oocysts per gram of feces. Cryptosporidium species in positive PCR products are determined by restriction fragment length polymorphism (RFLP) analysis using restriction enzymes SspI and MboII by DNA sequencing. Cryptosporidium parvum and Cryptosporidium ubiquitum are subtyped by nested-PCR-sequence analysis of the gp60 gene as previously described for calves [34].
DNA Sequence Analysis: To confirm the identification of Cryptosporidium ubiquitum and Cryptosporidium xiaoi, the secondary PCR products of the SSU rRNA gene from the two Cryptosporidium species are sequenced in both directions on an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The SSU rRNA gene products of Cryptosporidium parvum are not sequenced because it has a well-known SspI and MboII RFLP pattern. In addition, all PCR products of the gp60 gene are sequenced to identify Cryptosporidium parvum and Cryptosporidium ubiquitum subtypes. The generated sequences are assembled using the ChromasPro v.1.5 software (http:// www.technelysium.com.au/ChromasPro.html) and aligned with each other and with reference sequences (Ye et al., 2013).
Prevalence of Cryptosporidiosis in Lamb in Different Parts of Ethiopia
Table 3. Prevalence calculated from pooled published data of Cryptosporidium infection in sheep (especial focus on Ethiopia).
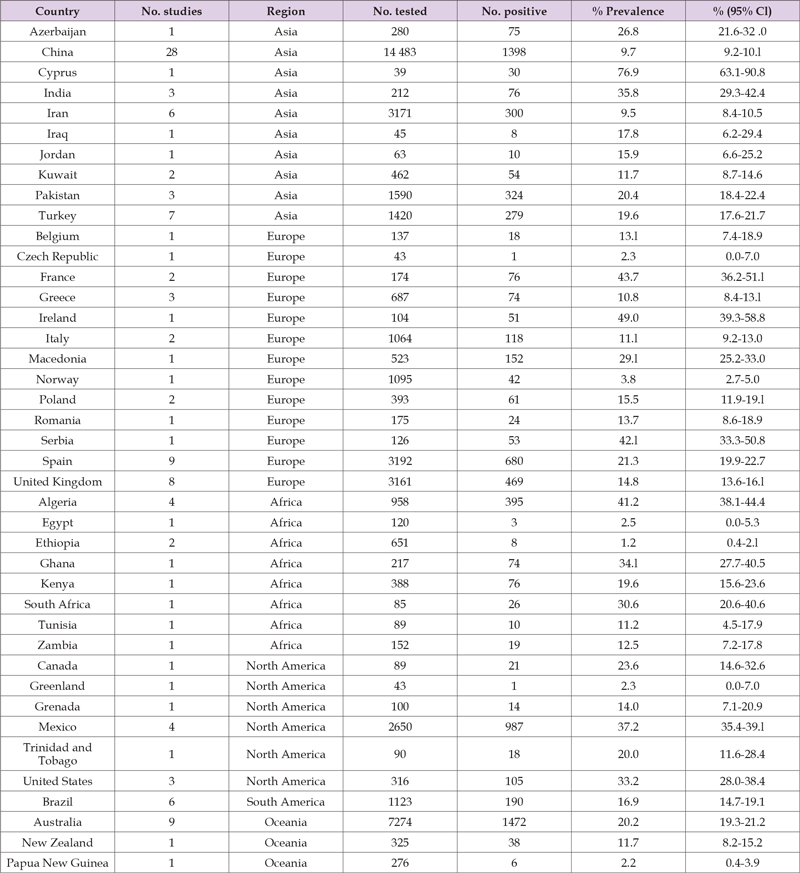
Note: Source: [41].
Cryptosporidiosis in Humans
Cryptosporidiosis is an emerging protozoan disease, caused by Cryptosporidium species, which can cause gastrointestinal infection in a wide variety of mammals including human worldwide. It causes watery diarrhea that can be severe after you get cryptosporidiosis from contaminated water, like pools or lakes, or from other people. It can cause life-threatening complications and become chronic if you have a weakened immune system [11,41]. Children between 1 and 4 years are the most likely to get cryptosporidiosis which is the second most common cause of diarrhea in children next to rotavirus infection. It spreads easily in kids because they don’t yet have good hand washing habits or understand how germs spread. Infected poop in diapers also helps spread crypto among young kids and their parents [42]. But also, one with over 75 years age, Live or work with young children, drink unfiltered or untreated water (often while traveling, hiking or camping), work with animals, particularly farm animals or livestock, frequent public pools or recreational water areas (lakes or rivers) and whom take care of someone who has cryptosporidiosis are at risk to get cryptosporidiosis easily [15]. In general if you’re living with a compromised immune system and get cryptosporidiosis, you’re at risk of ongoing and life-threatening illness. Half of all people living with AIDS will never get rid of Cryptosporidium once infected [2].
Parasitological Techniques in Human: Microscopic detection of Cryptosporidium species oocysts in fecal samples: After removal of the preservative through washing, the specimens were concentrated via formalin–ethyl acetate sedimentation, and a thin fecal smear was examined for each specimen after staining with modified Ziehl–Neelsen technique [43]. Briefly, slides were stained with carbol fuchsin and differentiated in 1% hydrochloric acid–alcohol (70%) for 1min before counterstaining with 1% methylene blue for 1 min. The stained slides were examined using an oil immersion lens at 100× magnification, where oocysts stained pink to red or deep purple against a blue background. The presence or absence of Cryptosporidium was recorded for each stool sample examined [14].
Molecular Techniques in Human:
DNA Extraction, Molecular Detection and Subtyping: Nucleic acid is extracted from all fecal specimens using for example QIAamp Power fecal DNA kit (Qiagen, France) following the manufacturer’s protocol. To enable the rapid detection and identification of Cryptosporidium hominis and Cryptosporidium parvum, two major species that are associated with human cryptosporidiosis, samples are screened using 18S ribosomal RNA (rRNA) nested PCR and real-time PCR as described elsewhere. Briefly, PCR is carried out in duplicate and consisted of two duplex reactions: a genus-specific PCR amplifying 300 bp of the Cryptosporidium 18S rRNA gene, duplexed
(i) With a Cryptosporidium parvum-specific PCR amplifying
166 bp of the LIB13 locus, and
(ii) With a Cryptosporidium hominis-specific PCR amplifying
169 bp of the LIB13 locus.
Thermocycling conditions are as follows: 95˚C for 10min, followed by 55 cycles of 95˚C for 15 s and 60˚C for 60s. Data are collected from each probe channel during each 60˚C annealing/extension phase. To correctly identify other species infecting human and to confirm results from the real-time PCR, genomic DNA extracts are subjected to a nested PCR-based sequencing protocol, targeting the 18S ribosomal RNA (rRNA) gene, as described elsewhere. For the primary PCR, the cycling protocol is as follows: 94˚C for 5 min; followed by 40 cycles of 94˚C for 30s, 58˚C for 45s, and 72˚C for 1min; with a final extension of 72˚C for 5min. For the secondary PCR, the protocol is as follows: 94˚C for 5min; followed by 40 cycles of 94˚C for 30s, 58˚C for 45s, and 72˚C for 45s; with a final extension of 72˚C for 5min. Products are visualized in 2% agarose gels using ethidium bromide staining. Positive samples are further subtyped by DNA sequencing of the GP60 gene. Subtyping is performed by sequencing a fragment of the GP60 gene. Each sample is amplified at least three times by nested PCR. Primers AL3531 and AL3533 are used in primary PCR and primers AL3532 and LX0029 are used in secondary PCR [26]. Reaction mixtures are prepared using 5 μL 10× DreamTaq Buffer, 0.2 mM of each deoxynucleoside triphosphate, 100 nM of each primer, 2.5 U DreamTaq polymerase, and 5 μL DNA template. Additionally, 1.25 μL of dimethyl sulfoxide is added to the mixture.
Cycle conditions are as follows: one cycle of 94˚C for 3 min; 39 cycles of a denaturation step at 94˚C for 45s, an annealing step at 54˚C (for both the first and the second rounds) for 45s, and an extension step at 72˚C for 1min; with a final extension for 10 min at 72˚C. Each amplification run included a negative control (PCR water) and two positive controls (genomic DNA from C. parvum oocysts purchased for example from INRAE Centre Val de Loire-Nouzilly France, and Cryptosporidium hominis genomic DNA from a fecal specimen collected at Rouen University Hospital). Products are visualized in 2% agarose gels using ethidium bromide staining and sequencing is used for identification and subtype confirmation. PCR amplicons are purified using exonuclease I/shrimp alkaline phosphatase (Exo-SAP-IT) (USB Corporation, Cleveland, Ohio, USA). They are sequenced in both directions using the same PCR primers at 3.2 uM in 10 μL reactions with Big Dye™ chemistry in an ABI 3500 sequence analyzer (Applied 229 Biosystems, California, and USA). Sequence chromatograms of each strand are examined with 4Peaks software and compared with published sequences in the Gene Bank data base using the Basic Local Alignment Search Tool (BLAST; www.ncbi.nlm.nih.gov/BLAST) [3].
Selected prevalence of Cryptosporidiosis in Humans in Ethiopia based on location.
Zoonosis can be broadly defined as diseases that are naturally transmissible between animals and people. Zoonosis are of concern for two main reasons: the health and economic burdens caused by zoonosis long known to cause disease in animals and people that persisting vulnerable groups and the potential for emerging zoonosis to give rise to novel disease outbreaks [44]. The burden of persisting zoonosis is mainly borne by poor people in developing countries, whilst emerging zoonosis are of more concern to the rich in developed countries with large economies and fewer other infectious disease problems [45]. More than 15% of the world’s population has no access to safe drinking water [46]. Waterborne parasitic protozoan diseases with worldwide distribution, result in four billion cases of diarrhoea, 1.6 million deaths annually (www.who.int) and 62.5 million Disability Adjusted Life Years (DALYs) worldwide [47,48]. Yet, despite the latest advances made in water treatment measures, protecting drinking water supplies against waterborne pathogens remains by far, as one of the most challenging concerns for the entire drinking water supply chain worldwide [46]. In response to this, in 2009, the World Health Organization has developed guidelines for water suppliers on how to implement “Water Safety Plans” (WSPs), in the hope of halving the number of people without safe access to drinking water by the end of 2015 [49].
In less developed countries, lack of basic infrastructure for providing safe drinking water is considered a major cause of poor water quality which contributes to the spread of endemic/epidemic waterborne diseases. However, even in industrialized nations, highly advanced infrastructures are not yet a protective factor against outbreaks [50]. This appears to be largely due to a lack of knowledge about the epidemiology and transmission dynamics of waterborne pathogens (example from animals ranging within the catchments) which leads to poor management practices for drinking water catchments [51]. Waterborne parasitic protozoans are responsible for the majority of waterborne outbreaks worldwide, with socio-economic impacts even in developed countries [47]. Of these, Cryptosporidium was the etiological agent in 60.3% (120) of the waterborne protozoan parasitic outbreaks that have been reported worldwide between 2004 and 2010. For the global water industry, therefore, Cryptosporidium represents the major public health concern, as its oocyst (the environmentally stable stage) is able to survive and penetrate routine wastewater treatment and is resistant to inactivation by commonly used drinking water disinfectants [52,16].
Current Status
Cryptosporidium is increasingly recognized as one of the major causes of moderate to severe diarrhea in developing countries. With treatment options limited, control relies on knowledge of the biology and transmission of the members of the genus responsible for disease. Currently, 26 species are recognized as valid on the basis of morphological, biological and molecular data. Of the nearly 20 Cryptosporidium species and genotypes that have been reported in humans, Cryptosporidium hominis and Cryptosporidium parvum are responsible for the majority of infections [53]. Livestock, particularly cattle, are one of the most important reservoirs of zoonotic infections. Domesticated and wild animals can each be infected with several Cryptosporidium species or genotypes that have only a narrow host range and therefore have no major public health significance. Recent advances in next-generation sequencing techniques will significantly improve our understanding of the taxonomy and transmission of Cryptosporidium species and the investigation of outbreaks and monitoring of emerging and virulent subtypes [54]. Important research gaps remain including a lack of subtyping tools for many Cryptosporidium species of public and veterinary health importance, and poor understanding of the genetic determinants of host specificity of Cryptosporidium species and impact of climate change on the transmission of Cryptosporidium [55].
Cryptosporidium species are well recognized as causes of diarrheal disease during waterborne epidemics and in immunocompromised hosts [56]. Studies have also drawn attention to an underestimated global burden and suggest major gaps in optimum diagnosis, treatment and immunization. Cryptosporidiosis is increasingly identified as an important cause of morbidity and mortality worldwide. Studies in low-resource settings and high-income countries have confirmed the importance of cryptosporidium as a cause of diarrhea and childhood malnutrition [57]. Diagnostic tests for cryptosporidium infection are suboptimum, necessitating specialized tests that are often insensitive [58]. Antigen-detection and PCR improve sensitivity and multiplexed antigen detection and molecular assays are underused. Therapy has some effect in healthy hosts and no proven efficacy in patients with AIDS [59]. Use of cryptosporidium genomes has helped to identify promising therapeutic targets and drugs are in development, but methods to assess the efficacy in-vitro and in animals are not well standardized. Partial immunity after exposure suggests the potential for successful vaccines and several are in development; however, surrogates of protection are not well defined [60,61]. Improved methods for propagation and genetic manipulation of the organism would be significant advances [41].
Cryptosporidium species distributes worldwide, especially in undeveloped and developing countries. The distribution has arisen when Cryptosporidium’s oocyst is defecated to water surface from human and animals (wildlife, domestic animals and livestock) through the feces. Both humans and animals had a high risk of infection from contaminated feed and water due to improper handling of manure on dairy farms and rural farmers’ households. The excreted oocysts are sustainable and tolerance with disinfectant, dilute bleach and chlorine. Transmission occurs when hosts expose with Cryptosporidium’s oocyst mainly by fecal-oral route through contaminated food and water or contact directly with animal feces and indirectly by cross contamination. Laboratory diagnosis of Cryptosporidium infection normally uses oocyst staining and examines under microscopy. However, these methods cannot differentiate morphology of each Cryptosporidium species, so immunological methods and molecular techniques are playing more roles for identification to species. Most of robust infected human and animals are asymptomatic and mild diarrhea, but violent symptoms will occur in the immune-compromised host. The best strategy to prevent and narrow down the spreading of this disease is keeping good personal hygiene in human coupled with reduction, control, or elimination the causative risk factors for other animals.
Based on the above conclusion, the following recommendations are forwarded:
A. To date, there has been no effective treatment or vaccination
for Cryptosporidium infection in animals hence maintaining
proper hygiene and management systems should be the first option
to prevent the transmission of infection in young calves and
lambs.
B. Since adult cattle and sheep excrete oocysts into the environment,
isolation of young calves and lambs from their dam is
important to decrease the possibility of acquiring infection from
their dams.
C. Proper wearing of protective gloves, clothes and shoes
should be important during handling animals to prevent the
transmission of Zoonotic Cryptosporidium species.
D. Dairy farmers and farmer households should handle, treat
and store animal manure properly in order to reduce the risk of
infection to both animals and humans.
E. Further investigation is necessary particularly on lambs in
West and South part of the country to have a national prevalence.
F. Finally national prevalence of Cryptosporidium infection in
animals and humans should have to be there to have effective
control and preventive program.
No competing interests existing.
AW: Conception of the review idea, drafting the manuscript and Structuring and edition. The author read and approved the final manuscript.


