Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
SG Borkar*
Received: January 19, 2022; Published: February 07, 2023
*Corresponding author: SG Borkar, Soil, Seed and Plant Disease Diagnostic Laboratory and Research centre, 103, Prestige Point Building, In front of Nashik Road Police Station, Nashik, Maharashtra state, 422 101, India
DOI: 10.26717/BJSTR.2023.48.007655
Health is a prime concern for the better and long life, which is affected by several factors like nutrition, immunity, infectious agents, mental status of a human etc. In recent years, it is demonstrated that all these factors influencing human health are governed by human gut microbes. Human Gut microbes are those microorganisms, including bacteria and archaea that lives in the gastro-intestinal (GI) tracts of humans. 95% of the body’s microbiota is found in the GI tract and plays a key role in digestive, metabolic, immune and neurological functions and are therefore very important for the gut and human health. These gut microorganisms perform a variety of useful functions , such as fermenting unused energy substrates, training the immune system, preventing the growth of harmful pathogenic species, regulating the development of the gut, producing vitamins (such as biotin and vitamin K) and hormones for the human body, development of enteric protection, metabolizing bile acids, sterols, xenobiotics, pharmaco-microbiomics and gut-brain axis. Apart from carbohydrates metabolism, gut microbiota can also metabolize other xenobiotics such as drugs, phytochemicals, and food toxicants. The dysregulation of the gut flora has been correlated with a variety of inflammatory and autoimmune conditions and obesity. Thus, human gut microbiota appears to play a major and significant role in maintaining the human health and disruption of this gut microbiota may lead to different diseased conditions. Factors that disrupt the gut microbiota population include antibiotics, stress, and parasites.
The human gut microbiota is dominated by four dominant bacterial phyla viz. Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria. Most bacteria belong to the Genera Bacteroides, Clostridium, Faecalibacterium, Eubacterium, Ruminococcus, Peptococcus, Peptostreptococcus, and Bifidobacterium. Other genera, such as Escherichia and Lactobacillus, are present to a lesser extent. Species from the genus Bacteroides alone constitute about 30% of all bacteria in the gut, suggesting that this genus is especially important in the functioning of the GI tract. About 99% of the large intestine flora are made up of obligate anaerobes such as Bacteroides and Bifidobacterium. Bifidobacterium and Lactobacillus genera (B. longum, B. breve, B. infantis; L. helveticus, L. rhamnosus, L. plantarum, and L. casei), had the most potential to be useful for certain central nervous system disorders. These GI tract microbes are also used as probiotics in treating the GI tract related medical conditions. These probiotics include the species of Bifidobacterium longum, B. breve, B. infantis, B. bifidum, B. adolescentis; Lactococcus cremoris, L. lactis; Enterococcus foeclum; Lactobacillus rhamnosus, L. acidophilus, L. casei, L. bulgaricus, L. gasseri, while the yeast species used as probiotics are Sacchoromyces boulardii and S. cerevisiae. During the treatment of different diseases with pharmaceutical drugs, there is considerable potential for interactions between drugs and an individual's microbiome, including: drugs altering the composition of the human microbiome, drug metabolism by microbial enzymes modifying the drug's pharmacokinetic profile, and microbial drug metabolism affecting a drug's clinical efficacy and toxicity profile. More than 30 drugs have been shown to be metabolized by gut microbiota. The microbial metabolism of drugs can sometimes inactivate the drug. Besides antibiotics the non-antibiotic drugs also impact the human gut-associated bacteria demonstrating that 24% of the drugs inhibited the growth of at least one of the gut bacterial strain. Thus, maintaining the equilibrium of the gut microbiota seems to be an essential aspect in modern human medicine in treating the human ailments. Besides, oral supplement of gut microbiota (probiotics), it can be advice to have a food having these gut microbes or probiotics. Probiotic bacteria with plant-based matrices (processed foods) are available, but there is lack of un-processed food/fresh food having these gut microbes. Although some of these gut microbes are associated with agricultural crop niches and systems, the idea to use them in the agricultural production is lacking far behind. The production of gut microbe’s assisted agricultural food may be useful to a larger extent, but no such program/protocol/research is being conducted in the agriculture sector world-over. The concept of gut microbiota assisted agricultural food production and its use may open a new technique in better management of human health.
Keywords: Human Health; Gut Microbes; Agriculture; Food; Nutrition; Probiotics
What are the human Gut Microbes
Human Gut microbiota or Gut microbes are the microorganisms, including bacteria and archaea, which lives in the gastro-intestinal (GI) tracts of humans (Moszak, et al. [1,2]). The GI tract is the tract or passage-way of the digestive system that leads from the mouth to the rectum. The GI tract contains all the major organs of the digestive system including the oesophagus, stomach, and intestine (Figure 1). Trillions of microorganism live inside the gut which comprises bacteria, viruses and non-pathogenic fungi. 95% of the body’s microbiota is found in the gut and are of good microbes. This microbiota plays a key role in digestive, metabolic, immune and neurological functions and are important for the gut and human health. Gut health can be defined as a state of well-being and absence of gastro-intestinal distress. It is determined by numerous factors and largely by the gut microbiota. The gut microbiota has broad impacts on human health, including effects on colonization of bad microbes, resistance to pathogens, maintaining the intestinal epithelium, metabolizing dietary and pharmaceutical compounds, controlling immune function, and even behaviour through the gut-brain axis. In humans, the gut flora is established at one to two years after birth, by which time the intestinal epithelium and the intestinal mucosal barrier that it secretes have co-developed in a way that, it is tolerant to and even supportive of, the gut flora and also provides a barrier to pathogenic organisms (Sommer, et al. [3,4]).
The microbial composition of the gut microbiota varies across the digestive tract. In the stomach and small intestine, relatively few species of bacteria are generally present (Guarner, et al. [5,6]). The colon, in contrast, contains the highest microbial density recorded in any habitat on Earth (Shapira [7]) with up to 1012 cells per gram of intestinal content (Guarner, et al. [5]). These bacteria represent between 300 and 1000 different species (Guarner, et al. [5,6]). However, 99% of the bacteria come from about 30 or 40 species (Beaugerie, et al. [8]). As a consequence of their abundance in the intestine, bacteria also make up to 60% of the dry mass of faeces (Stephen, et al. [9]). Fungi, protists, archaea, and viruses are also present in the gut flora, but less is known about their activities (Lozupone, et al. [10]). Over 99% of the bacteria in the gut are anaerobes, but in the cecum, aerobic bacteria reach high densities (Sherwood, et al. [11]). It is estimated that these gut flora have around a hundred times as many genes in total as there are in the human genome (Qin et al. 2010). Many species in the gut have not yet been studied outside of their habitat because most of these are not culturable (Sears, et al. [6,8]) (Shanahan, 2002). While there are a small number of core species of microbes shared by most individuals (Figure 2), populations of microbes can vary widely among different individuals (Tap, et al. [12]). Within an individual, microbe populations stay fairly constant over time, even though some alterations may occur with changes in lifestyle, diet and age (Guarner, et al. [5,13]).
The four dominant bacterial phyla in the human gut are Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria (Khanna, et al. [14]). Most bacteria belong to the generaBacteroides, Clostridium, Faecalibacterium,Eubacterium, Ruminococcus, Peptococcus, Peptostreptococcus, and Bifidobacterium (Guarner, et al. [5,8]) . Other genera, such as Escherichia and Lactobacillus, are present to a lesser extent (Guarner, et al. [5]). Species from the genus Bacteroides alone constitute about 30% of all bacteria in the gut, suggesting that this genus is especially important in the functioning of the GI tract (Sears, et al. [6]). Fungal genera that have been detected in the gut include Candida, Saccharomyces, Aspergillus, Penicillium, Rhodotorula, Trametes, Pleospora , Sclerotinia, Bullera, and Galactomyces, among others (Cui, et al. [15,16]). Rhodotorula is most frequently found in individuals with inflammatory bowel disease while Candida is most frequently found in individuals with hepatitis B cirrhosis and chronic hepatitis B (Cui, et al. [15])and are harmful gut microbes. Archaea constitute another large class of gut flora which are important in the metabolism of the bacterial products of fermentation. The relationship between some gut flora and humans is not merely commensal (a non-harmful coexistence), but rather a mutualistic relationship (Sherwood, et al. [11]). Some human gut microorganisms benefit the host by fermenting dietary fiber into short-chain fatty acids (SCFAs), such as acetic acid and butyric acid, which are then absorbed by the host (Quigley, et al. [17,18). Intestinal bacteria also play a role in metabolizing bile acids, sterols, and xenobiotics (Sherwood, et al. [11,18]).The systemic importance of the SCFAs and other compounds they produce are like hormones and the gut flora itself appears to function like an endocrine organ (Clarke, et al. [18]) and dysregulation of the gut flora has been correlated with a host of inflammatory and autoimmune conditions (Quigley, et al. [17,19]). These microorganisms perform a host of useful functions (Table 1), such as fermenting unused energy substrates, training the immune system via end products of metabolism like propionate and acetate, preventing growth of harmful species, regulating the development of the gut, producing vitamins for the host (such as biotin and vitamin K), and producing hormones to direct the host to store fats (Sherwood, et al. [11]). Extensive modification and imbalances of the gut microbiota and its microbiome or gene collection are associated with obesity (Ley [20]).
Gut Bacteriome
Bacteriome of Oral Cavity: The oral cavity contains Corynebacteria, Lactobacillus, Streptococcus and Staphylococcus.
Bacteriome of Stomach: Due to the high acidity of the stomach, most microorganisms cannot survive there. The main bacterial inhabitants of the stomach include Streptococcus, Staphylococcus, Lactobacillus, Peptostreptococcus (Sherwood, et al. [11]).
Bacteriome of Small Intestine: The small intestine contains a less numbers of microorganisms due to the proximity and influence of the stomach. Gram-positive cocci and rod-shaped bacteria are the predominant microorganisms found in the small intestine (Sherwood, et al. [11]). However, in the distal portion of the small intestine alkaline conditions support gram-negative bacteria of the Enterobacteriaceae (Sherwood, et al. [11]). The bacterial flora of the small intestine aid in a wide range of intestinal functions. The bacterial flora provide regulatory signals that enable the development and utility of the gut. Overgrowth of bacteria in the small intestine can lead to intestinal failure (Quigley, et al. [21]). The anaerobic bacterial genera include Bifidobacterium, Clostridium and Bacteroides while the aerobic bacteria genera include Escherichia, Enterococcus and Streptococcus.
Bacteriome of Colon: The bacteria found in colon comprise of different bacterial species (Table 2). In addition, the large intestine contains the largest bacterial ecosystem in the human body (Sherwood, et al. [11]). About 99% of the large intestine and faeces flora are made up of obligate anaerobes such as Bacteroides and Bifidobacterium (Adams, et al. [22]). Factors that disrupt the microorganism population of the large intestine include antibiotics, stress, and parasites (Sherwood, et al. [11]).
The Bacterial Flora in the Gut
The bacterial flora in the gut comprises both good and bad bacterial flora (Table 3).
Type of Good Bacterial Species in Gut:
Bifidobacteria and Lactobacilli Species: Human cannot produce most vitamins, and must therefore inject them through their diet. Bifidobacteria, Lactobacilli and other commensal bacteria can synthesize vitamins de novo. These vitamins includes Folate (vitamin B9), Riboflavin (vitamin B2), Cobalamin (vitamin B12) etc. Bifidobacteria also alter the activity of dendritic cell, which leads to a more efficacious anti-tumour response. The habitat of these bacteria is small intestine and colon.
Bacteroides
Bacteroides Fragilis: This bacterial species produces polysaccharide A (PSA). PSA is recognized by dendritic cell, and presented to CD4 + T cells of the immune system. Upon presentation of PSA, the CD4 + T cells differentiate in to regulatory T cells, which reduce inflammation. fragilis also increases the efficacy of anti-CTLA-4 therapy, which is used to treat cancer.
Bacteroides Thetaiotaomicron: This bacterial species produces enzymes that can digest complex plant sugars that human enzyme are unable to digest. This allows human to gain energy from otherwise indigestible plant sugar (10% of our daily absorbed calories come from these plant sugar). The habitat of these bacterial species is small intestine and colon.
Clostridium Species: These bacterial species produce butyrate, while facilitate differentiation of CD4 + T cells in to regulatory T cells and stimulates immune cells to produce anti-inflammatory cytokines. Promotes intestinal homeostasis by preventing inflammation. This bacteria convert primary bile acids into secondary bile acid (e.g. deoxycholic acid) which inhibits the growth of bad bacteria particularly difficile. The habitat of these bacterial species is small intestine and colon. Intake of Fruits and vegetables in the diet promotes growth of good Clostridium species.
Types of Bad Bacterial Species in Gut:
Clostridium Difficile: These are available in colon and causes watery diarrhea. In the absence of good bacteria which are killed due to antibiotic treatment, difficile pose a threat.
Eggerthella Lenta: These bacteria are present in colon and implicated as a cause of ulcerative colitis, liver and renal abscesses and systemic bacteremia. These can inactivate “digoxin”, a drug used to treat heart conditions (e.g. atrial fibrillation). These are responsible for brain and liver abscesses, heart disease.
Desulfovibrio Desulfuricans (and other Proteobacteria): These are present in the colon. These bacteria produces Trimethylamine (TMA), which is converted by the liver into Trimethylamine N-oxide (TMAO). High TMAO is linked to a higher risk of cardiovascular disease, blood clots and atherosclerosis and is thus responsible for Heart disease, cholecystitis, and abscesses.
Proteobacteria Species: These bacteria are present in small intestine and colon. These can convert tryptophan into indole, which is converted to indoxyl sulfate in the liver. The indoxyl sulfate contributes to uremic toxicity. These are responsible for renal disease and interstitial fibrosis.
Escherichia Coli (Esp Strain NC101 & Nissle 1917): This is available in the colon. Up to 34% of commensal E.coli strains isolated from human produce colibactin, which induces breaks in the DNA of human cells. This bacterial species induce IBD and colorectal cancer. The composition of human gut microbiota changes over time, and is influenced by various factors, and as overall health changes (Quigley, et al. [17,19]).
Factors Influencing Gut Microbiota in Human
Age: It has been demonstrated that there are common patterns of microbiome composition evolution during life (Gerritsen, et al. [23]). In general, the diversity of microbiota composition of faecal samples is significantly higher in adults than in children, although interpersonal differences are higher in children than in adults (Yatsunenko, et al. [24]). Much of the maturation of microbiota into an adult-like configuration happens during the three first years of life (Yatsunenko, et al. [24]). As the microbiome composition changes, so does the composition of bacterial proteins produced in the gut. In adult microbiomes, a high prevalence of enzymes involved in fermentation, methanogenesis and the metabolism of arginine, glutamate, aspartate and lysine have been found. In contrast, in infant microbiomes the dominant enzymes are involved in cysteine metabolism and fermentation pathways (Yatsunenko, et al. [24]).
Diet: Studies and statistical analyses have identified the different bacterial genera in gut microbiota and their associations with nutrient intake. Gut microflora is mainly composed of three enterotypes: Prevotella, Bacteroides, and Ruminococcus. There is an association between the concentration of each microbial community and diet. For example, Prevotella is related to carbohydrates and simple sugars, while Bacteroides is associated with proteins, amino acids, and saturated fats. Specialist microbes that break down mucin survive on their host's carbohydrate excretions (Alcock, et al. [25]). One enterotype will dominate depending on the diet. Altering the diet will result in a corresponding change in the numbers of species (Wu, et al. [26]). A 2021 study suggests that childhood diet and exercise can substantially affect adult microbiome composition and diversity. Further studies have indicated a large difference in the composition of microbiota between European and rural African children. The faecal bacteria of children from Florence were compared to that of children from the small rural village of Boulpon in Burkina Faso. The diet of a typical child living in this village is largely lacking in fats and animal proteins and rich in polysaccharides and plant proteins. The faecal bacteria of European children were dominated by Firmicutes and showed a marked reduction in biodiversity, while the faecal bacteria of the Boulpon children was dominated by Bacteroidetes. The increased biodiversity and different composition of gut flora in African populations may aid in the digestion of normally indigestible plant polysaccharides and also may result in a reduced incidence of non-infectious colonic diseases (De Filippo, et al. [27]). Malnourished children have less mature and less diverse gut microbiota than healthy children, and changes in the microbiome associated with nutrient scarcity can in turn be a pathophysiological cause of malnutrition (Jonkers, et al. [28,29]). Malnourished children also typically have more potentially pathogenic gut flora, and more yeast in their mouths and throats (Rytter, et al. [30]). Altering diet may lead to changes in gut microbiota composition and diversity (Alcock, et al. [25]).
Socio-Economic Status: As of 2020, at least two studies have demonstrated a link between an individual's socio-economic status (SES) and their gut microbiota. A study in Chicago found that individuals in higher SES neighbour-hoods had greater microbiota diversity. People from higher SES neighbour-hoods also had more abundant Bacteroides bacteria. Similarly, a study of twins in the United Kingdom found that higher SES was also linked with a greater gut diversity (Renson, et al. [31]).
Acquisition of Gut Microbiota in Human
The establishment of a gut microbiota is crucial to the health of an adult, as well as the functioning of the gastro-intestinal tract (Turroni, et al. [32]). In humans, a gut microbiota similar to an adult's is formed within one to two years of birth as microbiota are acquired through parent-to-child transmission and transfer from food, water, and other environmental sources (Davenport, et al. [33,3]). The traditional view of the gastro-intestinal tract of a normal foetus is that it is sterile, although this view has been challenged in the past few years (Perez-Munoz, et al. [34]). Multiple lines of evidence have begun to emerge that suggest there may be bacteria in the intrauterine environment. In humans, research has shown that microbial colonization may occur in the foetus (Matamoros, et al. [35]) with one study showing Lactobacillus and Bifidobacterium species were present in placental biopsies (Mueller et al. [36]). During birth and rapidly thereafter, bacteria from the mother and the surrounding environment colonize the infant's gut (Sommer, et al. [3]). During the year of life, the composition of the gut flora is generally simple and changes a great deal with time and is not the same across individuals (Sommer, et al. [3]).
The initial bacterial population are generally facultative anaerobic organisms; investigators believe that these initial colonizers decrease the oxygen concentration in the gut, which in turn allows obligate anaerobic bacteria like Bacteroides, Actinobacteria, and Firmicutes to become established and thrive (Sommer, et al. [3]). Breast-fed babies become dominated by bifidobacteria, possibly due to the contents of bifidobacterial growth factors in breast milk, and by the fact that breast milk carries prebiotic components, allowing for healthy bacterial growth (Mueller, et al. [36,37]). In contrast, the microbiota of formula-fed infants is more diverse, with high numbers of Enterobacteriaceae, enterococci, bifidobacteria, Bacteroides, and clostridia (Fanaro, et al. [38]). Caesarean section, antibiotics, and formula feeding may alter the gut microbiome composition (Mueller et al. [36]). Children treated with antibiotics have less stable, and less diverse microbial communities (Yassour, et al. [39]). Caesarean sections have been shown to be disruptive to mother-offspring transmission of bacteria, which impacts the overall health of the offspring by raising risks of disease such as celiacs, asthma, and type 1 diabetes (Mueller, et al. [36]). This further evidences the importance of a healthy gut microbiome. Various methods of microbiome restoration are being explored, typically involving exposing the infant to maternal vaginal contents, and oral probiotics (Mueller, et al. [36]).
What are the Important Functions of Gut Microbiota
When the study of gut flora began in 1995, (Gibson, et al. [40]) it was thought to have three key roles (Table 4): direct defence against pathogens, fortification of host defence by its role in developing and maintaining the intestinal epithelium and inducing antibody production there, and metabolizing otherwise indigestible compounds in food. Subsequent work discovered its role in training the developing immune system, and yet further work focused on its role in the gut-brain axis (Wang, et al. [41]).
Direct Inhibition of Pathogens: The gut flora community plays a direct role in defending against pathogens by fully colonising the space, making use of all available nutrients, and by secreting compounds that kill or inhibit unwelcome organisms that would compete for nutrients with it. These compounds are known as cytokines (Yoon, et al. [42]). Different strains of gut bacteria cause the production of different cytokines. Cytokines are chemical compounds produced by our immune system for initiating the inflammatory response against infections. Disruption of the gut flora allows competing organisms like Clostridium difficile to become established that otherwise are kept in abeyance (Yoon, et al. [42]).
Development of Enteric Protection and Immune System: In humans, a gut flora similar to an adult's is formed within one to two years of birth (Sommer, et al. [3]). As the gut flora gets established, the lining of the intestines – the intestinal epithelium and the intestinal mucosal barrier that it secretes – develop as well, in a way that is tolerant to, and even supportive of, commensalistic microorganisms to a certain extent and also provides a barrier to pathogenic ones (Sommer, et al. [3]). Specifically, goblet cells that produce the mucosa proliferate, and the mucosa layer thickens, providing an outside mucosal layer in which "friendly" microorganisms can anchor and feed, and an inner layer that even these organisms cannot penetrate (Sommer, et al. [3,4]). Additionally, the development of gut-associated lymphoid tissue (GALT), which forms part of the intestinal epithelium and which detects and reacts to pathogens, appears and develops during the time that the gut flora develops and established (Sommer, et al. [3]). The GALT that develops is tolerant to gut flora species, but not to other microorganisms (Sommer, et al. [3]). GALT also normally becomes tolerant to food to which the infant is exposed, as well as digestive products of food, and gut flora's metabolites (molecules formed from metabolism) produced from food (Sommer, et al. [3]). The human immune system creates cytokines that can drive the immune system to produce inflammation in order to protect itself, and that can tamp down the immune response to maintain homeostasis and allow healing after insult or injury (Sommer, et al. [3]).
Different bacterial species that appear in gut flora have been shown to be able to drive the immune system to create cytokines selectively; for example Bacteroides fragilis and some Clostridia species appear to drive an anti-inflammatory response, while some segmented filamentous bacteria drive the production of inflammatory cytokines (Sommer, et al. [3,43]). Gut flora can also regulate the production of antibodies by the immune system (Sommer, et al. [3,44]). One function of this regulation is to cause B cells to class switch to IgA. In most cases B cells need activation from T helper cells to induce class switching; however, in another pathway, gut flora cause NF-kB signalling by intestinal epithelial cells which results in further signalling molecules being secreted (Peterson, et al. [45]). These signalling molecules interact with B cells to induce class switching to IgA (Peterson, et al. [45]). IgA is an important type of antibody that is used in mucosal environments like the gut. It has been shown that IgA can help diversify the gut community and helps in getting rid of bacteria that cause inflammatory responses (Honda, et al. [46]). Ultimately, IgA maintains a healthy environment between the host and gut bacteria (Honda, et al. [46]). These cytokines and antibodies can have effects outside the gut, in the lungs and other tissues (Sommer, et al. [3]). The immune system can also be altered due to the gut bacteria's ability to produce metabolites that can affect cells in the immune system. For example short-chain fatty acids (SCFA) can be produced by some gut bacteria through fermentation (Levy, et al. [47]). SCFAs stimulate a rapid increase in the production of innate immune cells like neutrophils, basophils and eosinophils (Levy, et al. [47]). These cells are part of the innate immune system that try to limit the spread of infection.
Metabolism: The biosynthesis of bioactive compounds (indole and certain other derivatives) from tryptophan is carried out by bacteria in the gut (Zhang, et al. [48]). Indole is produced from tryptophan by bacteria that express tryptophanase (Zhang, et al. [48]). Clostridium sporogenes metabolizes tryptophan into indole and subsequently 3-indolepropionic acid (IPA), (Wikoff, et al. [49]) a highly potent neuroprotective antioxidant that scavenges hydroxyl radicals (Zhang, et al. [48,50,51]). IPA binds to the pregnane X receptor (PXR) in intestinal cells, thereby facilitating mucosal homeostasis and barrier function (Zhang, et al. [48]). Following absorption from the intestine and distribution to the brain, IPA confers a neuroprotective effect against cerebral ischemia and Alzheimer's disease (Zhang, et al. [48]). Lactobacillus species metabolize tryptophan into indole-3-aldehyde (I3A) which acts on the aryl hydrocarbon receptor (AhR) in intestinal immune cells, in turn increasing interleukin-22 (IL-22) production (Zhang and Davis, 2016). Indole itself triggers the secretion of glucagon-like peptide-1 (GLP-1) in intestinal L cells and acts as a ligand for AhR (Zhang, et al. [48]). Indole can also be metabolized by the liver into indoxyl sulfate, a compound that is toxic in high concentrations and associated with vascular disease and renal dysfunction (Zhang, et al. [48]). AST-120 (activated charcoal), an intestinal sorbent that is taken by mouth, adsorbs indole, in turn decreasing the concentration of indoxyl sulfate in blood plasma (Zhang, et al. [48]). Without gut flora, the human body would be unable to utilize some of the undigested carbohydrates it consumes, because some types of gut flora have enzymes that human cells lack for breaking down certain polysaccharides (Clarke, et al. [18). Carbohydrates that humans cannot digest without bacterial help include certain starches, fiber, oligosaccharides, and sugars that the body failed to digest and absorb like lactose in the case of lactose intolerance and sugar alcohols, mucus produced by the gut, and proteins (Quigley, et al. [17,18]).
Bacteria turn carbohydrates they ferment into short-chain fatty acids by a form of fermentation called saccharolytic fermentation (Gibson, et al. [52]). Products include acetic acid, propionic acid and butyric acid (Beaugeric, et al. [8,52]). These materials can be used by host cells, providing a major source of energy and nutrients (Gibson, et al. [52]). Gases (which are involved in signalling (Hoppor, et al. [53]) and may cause flatulence and organic acids, such as lactic acid, are also produced by fermentation (Beaugeric, et al. [8]). Acetic acid is used by muscle, propionic acid facilitates liver production of ATP, and butyric acid provides energy to gut cells (Gibson, et al. [52]). Gut microbiota also synthesize vitamins like biotin and folate, and facilitate absorption of dietary minerals, including magnesium, calcium, and iron (Guarner, et al. [5,13]). Methanobrevibacter smithii is unique because it is not a species of bacteria, but rather a member of domain Archaea, and is the most abundant methane-producing archaeal species in the human gastrointestinal microbiota (Rajilic-stojanovic, et al. [54]). Gut microbiota also serve as a source of Vitamins K and B12 that are not produced by the body or produced in little amount (Hill, et al. [55,56]).
Pharmaco-Microbiomics: The human metagenome (i.e., the genetic composition of an individual and all microorganisms that reside on or within the individual's body) varies considerably between individuals (El-Rakaiby, et al. [57,58]), since the total number of microbial and viral cells in the human body (over 100 trillion) greatly out numbers Homo sapiens cells (tens of trillions) (El-Rakaiby, et al. [57,59]). There is considerable potential for interactions between drugs and an individual's microbiome, including: drugs altering the composition of the human microbiome, drug metabolism by microbial enzymes modifying the drug's pharmacokinetic profile, and microbial drug metabolism affecting a drug's clinical efficacy and toxicity profile (El-Rakaiby, et al. [57,58,60]). Apart from carbohydrates, gut microbiota can also metabolize other xenobiotics such as drugs, phytochemicals, and food toxicants. More than 30 drugs have been shown to be metabolized by gut microbiota (Sousa, et al. [61]). The microbial metabolism of drugs can sometimes inactivate the drug (Haiser, et.al. [62]).
Gut-Brain Axis: The gut-brain axis is the biochemical signalling that takes place between the gastrointestinal tract and the central nervous system (Wang, et al. [14]). That term has been expanded to include the role of the gut microbiota in the interplay; the term "microbiome-gut-brain axis" is sometimes used to describe paradigms explicitly including the gut microbiota (Wang, et al. [14,63,64]). Broadly defined, the gut-brain axis includes the central nervous system, neuro-endocrine and neuro-immune systems including the hypothalamic–pituitary–adrenal axis (HPA axis), sympathetic and parasympathetic arms of the autonomic nervous system including the enteric nervous system, the vagus nerve, and the gut microbiota (Wang, et al. [14,63]). A systematic review from 2016 examined the preclinical and small human trials that have been conducted with certain commercially available strains of probiotic bacteria and found that among those tested, Bifidobacterium and Lactobacillus genera ( longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei), had the most potential to be useful for certain central nervous system disorders (Wang, et al. [65]).
Factors Responsible for Alterations in Microbiota Balance
Effects of Antibiotic Use: Altering the numbers of gut bacteria, for example by taking broad-spectrum antibiotics, may affect the host's health and ability to digest food (Carman, et al. [66]). Antibiotics can cause antibiotic-associated diarrhea by irritating the bowel directly, changing the levels of microbiota, or allowing pathogenic bacteria to grow (Beaugeric, et al. [8]). Another harmful effect of antibiotics is the increase in numbers of antibiotic-resistant bacteria found after their use, which, when they invade the host, cause illnesses that are difficult to treat with antibiotics (Carman, et al. [66]). Changing the numbers and species of gut microbiota can reduce the body's ability to ferment carbohydrates and metabolize bile acids and may cause diarrhea. Carbohydrates that are not broken down may absorb too much water and cause runny stools, or lack of SCFAs produced by gut microbiota could cause diarrhea (Beaugeric, et al. [8]). A reduction in levels of native bacterial species also disrupts their ability to inhibit the growth of harmful species such as difficile and Salmonella kedougou, and these species can get out of hand, though their over growth may be incidental and not be the true cause of diarrhea (Guarner, et al. [5,8,66]).
Emerging treatment protocols for C. difficile infections involve fecal microbiota transplantation of donor feces (Hyas et.al, 2020). Initial reports of treatment describe success rates of 90%, with few side effects. Efficacy is speculated to result from restoring bacterial balances of bacteroides and firmicutes classes of bacteria (Brandt, et al. [67). The composition of the gut microbiome also changes in severe illnesses, due not only to antibiotic use but also to such factors as ischemia of the gut, failure to eat, and immune compromise. Negative effects from this have led to interest in selective digestive tract decontamination, a treatment to kill only pathogenic bacteria and allow the re-establishment of healthy ones (Knight, et al. [68]). Antibiotics alter the population of the microbiota in the gastrointestinal tract, and this may change the intra-community metabolic interactions, modify caloric intake by using carbohydrates, and globally affects host metabolic, hormonal and immune homeostasis (Cho, et al. [69]). There is reasonable evidence that taking probiotics containing Lactobacillus species may help prevent antibiotic-associated diarrhea and that taking probiotics with Saccharomyces (e.g., Saccharomyces boulardii) may help to prevent Clostridium difficile infection following systemic antibiotic treatment (Schneiderhan, et al. [70]).
Pregnancy: The gut microbiota of a woman changes as pregnancy advances, with the changes similar to those seen in metabolic syndromes such as diabetes. The change in gut microbiota causes no ill effects. The new-born's gut microbiota resemble the mother's first-trimester samples. The diversity of the microbiome decreases from the first to third trimester, as the numbers of certain species go up (Mueller, et al. [36,71]).
Role of Bad Gut Bacteria in Causing Diseases
Proliferation of bad Gut Bacteria in the digestive tract can contribute to and be affected by disease in various ways. The presence or over-abundance of some kinds of bacteria may contribute to inflammatory disorders such as inflammatory bowel disease (Guarner, et al. [5]). Additionally, metabolites from certain members of the gut flora may influence host signalling pathways, contributing to disorders such as obesity and colon cancer (Guarner, et al. [5]). Alternatively, in the event of a breakdown of the gut epithelium, the intrusion of gut flora components into other host compartments can lead to sepsis (Guarner, et al. [5]). Following major health issues are associated with bad Gut microbes.
Ulcers: Helicobacter pylori infection can initiate formation of stomach ulcers when the bacteria penetrate the stomach epithelial lining, then causing an inflammatory phagocytotic response (Kamboj, et al. [72]). In turn, the inflammation damages parietal cells which release excessive hydrochloric acid into the stomach and produce less of the protective mucus (Anonymous [73). Injury to the stomach lining, leading to ulcers, develops when gastric acid overwhelms the defensive properties of cells and inhibits endogenous prostaglandin synthesis, reduces mucus and bicarbonate secretion, reduces mucosal blood flow, and lowers resistance to injury (Anonymous [73). Reduced protective properties of the stomach lining increase vulnerability to further injury and ulcer formation by stomach acid, pepsin, and bile salts (Kamboj, et al. [72,73]).
Bowel Perforation: Normally-commensal bacteria can harm the host if they extrude from the intestinal tract (Sommer, et al. [3,4]). Translocation, which occurs when bacteria leave the gut through its mucosal lining, can occur in a number of different diseases (Faderl, et al. [4]). If the gut is perforated, bacteria invade the interstitium, causing a potentially fatal infection (Sherwood, et al. [11]).
Inflammatory Bowel Diseases (IBD): The two main types of inflammatory bowel diseases, Crohn's disease and ulcerative colitis, are chronic inflammatory disorders of the gut; the causes of these diseases are unknown and issues with the gut flora and its relationship with the host have been implicated in these conditions (Shen, et al. [19,74-76]). Additionally, it appears that interactions of gut flora with the gut-brain axis have a role in IBD, with physiological stress mediated through the hypothalamic–pituitary–adrenal axis driving changes to intestinal epithelium and the gut flora in turn releasing factors and metabolites that trigger signalling in the enteric nervous system and the vagus nerve (Saxena, et al. [77]). The diversity of gut flora appears to be significantly diminished in people with inflammatory bowel diseases compared to healthy people. Additionally, in people with ulcerative colitis, Proteobacteria and Actinobacteria appear to dominate while in people with Crohn's disease, Enterococcus faecium and several Proteobacteria appear to be over-represented (Saxena, et al. [77]). There is reasonable evidence that correcting gut flora imbalances by taking probiotics with Lactobacilli and Bifidobacteria can reduce visceral pain and gut inflammation in IBD (Schneiderhan, et al. [70]).
Irritable Bowel Syndrome: Irritable bowel syndrome is a result of stress and chronic activation of the HPA axis which causes the symptoms like abdominal pain, changes in bowel movements, and an increase in pro-inflammatory cytokines. Overall, studies have found that the luminal and mucosal microbiota are changed in irritable bowel syndrome individuals, and these changes can relate to the type of irritation such as diarrhea or constipation. Also, there is a decrease in the diversity of the microbiome with low levels of fecal Lactobacilli and Bifidobacteria; high levels of facultative anaerobic bacteria such as Escherichia coli, and increased ratios of Firmicutes: Bacteroidetes (Dinan, et al. [64]).
Other Inflammatory or Autoimmune Conditions: Allergy, asthma, and diabetes mellitus are autoimmune and inflammatory disorders of unknown cause, but have been linked to imbalances in the gut flora and its relationship with the host (Shen, et al. [19]). As of 2016 it was not clear if changes to the gut flora cause these auto-immune and inflammatory disorders or are a product of or adaptation to them (Shen, et al. [19,78]).
Asthma: With asthma, two hypotheses have been posed to explain its rising prevalence in the developed world. The hygiene hypothesis posits that children in the developed world are not exposed to enough microbes and thus may contain lower prevalence of specific bacterial taxa that play protective roles (Arrieta, et al. [79]). The second hypothesis focuses on the Western pattern diet, which lacks whole grains and fiber and has an over-abundance of simple sugars (Shen, et al. [19]). Both hypotheses converge on the role of short-chain fatty acids (SCFAs) in immuno-modulation. These bacterial fermentation metabolites are involved in immune signalling that prevents the triggering of asthma and lower SCFA levels are associated with the disease (Arrieta, et al. [79,80]). Lacking protective genera such as Lachnospira, Veillonella, Rothia and Faecalibacterium has been linked to reduced SCFA levels (Arrieta et.al, 2015). Further, SCFAs are the product of bacterial fermentation of fiber, which is low in the Western pattern diet (Shen, et al. [19,80]). SCFAs offer a link between gut flora and immune disorders, and as of 2016, this was an active area of research. Similar hypotheses have also been posited for the rise of food and other allergies (Ipci, et al. [81]).
Diabetes Mellitus Type 1: The connection between the gut microbiota and diabetes mellitus type 1 has also been linked to SCFAs, such as butyrate and acetate. Diets yielding butyrate and acetate from bacterial fermentation show increased Treg expression (Marino, et al. [82]). Treg cells down-regulate effector T cells, which in turn reduces the inflammatory response in the gut (Bettelli, et al. [83]). Butyrate is an energy source for colon cells. Butyrate-yielding diets thus decrease gut permeability by providing sufficient energy for the formation of tight junctions (Saemann, et al. [84]). Additionally, butyrate has also been shown to decrease insulin resistance, suggesting gut communities low in butyrate-producing microbes may increase chances of acquiring diabetes mellitus type 2 (Gao et.al, 2009). Butyrate-yielding diets may also have potential colorectal cancer suppression effects (Saemann, et al. [84]).
Obesity and Metabolic Syndrome: The gut flora has also been implicated in obesity and metabolic syndrome due to the key role it plays in the digestive process. The Western pattern diet appears to drive and maintain changes in the gut flora that in turn change how much energy is derived from food and how that energy is used (Boulange, et al. [76,85]). One aspect of a healthy diet that is often lacking in the Western-pattern diet is fiber and other complex carbohydrates that a healthy gut flora require flourishing. The changes to gut flora in response to a Western-pattern diet appear to increase the amount of energy generated by the gut flora which may contribute to obesity and metabolic syndrome (Schneiderhan, et al. [70]). There is also evidence that microbiota influence eating behaviours based on the preferences of the microbiota, which can lead to the host consuming more food eventually resulting in obesity. It has generally been observed that with higher gut microbiome diversity, the microbiota will spend energy and resources on competing with other microbiota and less on manipulating the host.
The opposite is seen with lower gut microbiome diversity, and these microbiotas may work together to create host food cravings (Alcock, et al. [25]). Additionally, the liver plays a dominant role in blood glucose homeostasis by maintaining a balance between the uptake and storage of glucose through the metabolic pathways of glycogenesis and gluconeogenesis. Intestinal lipids regulate glucose homeostasis involving a gut-brain-liver axis. The direct administration of lipids into the upper intestine increases the long chain fatty acyl-coenzyme A (LCFA-CoA) levels in the upper intestines and suppresses glucose production even under sub-diaphragmatic vagotomy or gut vagal differentiation. This interrupts the neural connection between the brain and the gut and blocks the upper intestinal lipids' ability to inhibit glucose production. The gut-brain-liver axis and gut microbiota composition can regulate the glucose homeostasis in the liver and provide potential therapeutic methods to treat obesity and diabetes (Chen, et al. [86]). Just as gut flora can function in a feedback loop that can drive the development of obesity, there is evidence that restricting intake of calories (i.e., dieting) can drive changes to the composition of the gut flora (Boulange, et al. [76]).
Liver Disease: As the liver is fed directly by the portal vein, whatever crosses the intestinal epithelium and the intestinal mucosal barrier enters the liver, as do cytokines generated there (Minemura, et al. [87]). Dysbiosis in the gut flora has been linked with the development of cirrhosis and non-alcoholic fatty liver disease (Minemura, et al. [87]).
Cancer: Some genera of bacteria, such as Bacteroides and Clostridium, have been associated with an increase in tumor growth rate, while other genera, such as Lactobacillus and Bifidobacteria, are known to prevent tumor formation (Guarner, et al. [5]). As of December 2017 there was preliminary and indirect evidence that gut microbiota might mediate response to PD-1 inhibitors; the mechanism was unknown (Syn, et al. [88]).
Neuro-Psychiatric: Interest in the relationship between gut flora and neuro-psychiatric issues was sparked by a 2014 study showing that germ-free mice showed an exaggerated HPA axis response to stress compared to non-GF laboratory mice (Wang, et al. [41]). As of January 2016, most of the work that has been done on the role of gut flora in the gut-brain axis had been conducted in animals, or characterizing the various neuroactive compounds that gut flora can produce, and studies with humans measuring differences between people with various psychiatric and neurological differences, or changes to gut flora in response to stress, or measuring effects of various probiotics (dubbed "psychobiotics in this context), had generally been small and could not be generalized; whether changes to gut flora are a result of disease, a cause of disease, or both in any number of possible feedback loops in the gut-brain axis, remained unclear (Wang, et al. [41,70]). systematic review from 2016 examined the preclinical and small human trials that have been conducted with certain commercially available strains of probiotic bacteria and found that among those tested, the genera Bifidobacterium and Lactobacillus ( longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei) had the most potential to be useful for certain central nervous system disorders (Wang, et al. [65]).
Effect of Non-Antibiotic Drug on Gut Microbe
Tests for whether non-antibiotic drugs may impact human gut-associated bacteria were performed by in vitro analysis on more than 1000 marketed drugs against 40 gut bacterial strains, demonstrating that 24% of the drugs inhibited the growth of at least one of the bacterial strains (Maier, et al. [89]).
Probiotics, Prebiotics, Synbiotics, and Pharma-Biotics
Probiotics are microorganisms that are believed to provide health benefits when consumed (Hill, et al. [90,91]). while prebiotics are typically non-digestible, fiber compounds that pass undigested through the upper part of the gastrointestinal tract and stimulate the growth or activity of advantageous gut flora by acting as substrate for them (Gibson, et al. [52,92]). Synbiotics refers to food ingredients or dietary supplements combining probiotics and prebiotics in a form of synergism (Pandey, et al. [93]). The term "pharma-biotics" is used in various ways, to mean: pharmaceutical formulations (standardized manufacturing that can obtain regulatory approval as a drug) of probiotics, prebiotics, or synbiotics; (Broecky, et al. [94]) probiotics that have been genetically engineered or otherwise optimized for best performance (shelf life, survival in the digestive tract, etc.); (Sleator, et al. [95]) and the natural products of gut flora metabolism (vitamins, etc.). (Patterson, et al. [96]). There is some evidence that treatment with some probiotic strains of bacteria may be effective in irritable bowel syndrome and chronic idiopathic constipation. Those organisms most likely to result in a decrease of symptoms have included Enterococcus faecium, Lactobacillus plantarum, Lactobacillus rhamnosus, Propionibacterium freudenreichii, Bifidobacterium breve, Lactobacillus reuteri, Lactobacillus salivarius, Bifidobacterium infantis and Streptococcus thermophilus (Ford, et al. [97-99]).
Bacteria And Yeast Used as Probiotics: The bacterial species used as probiotics are Bifidobacterium longum, B. breve, B. infantis, B. bifidum, B. adolescentis, Lactococcus cremoris, L. lactis, Enterococcus foeclum, Lactobacillus rhamnosus, L. acidophilus, L. casei, L. bulgaricus, L. gasseri, while the yeast species used as probiotics are Sacchoromyces boulardii and cerevisiae.
Present Status of Natural Association of Human Gut Microbes With/In Agricultural Crops
Natural association of some of the gut microbes with crop plant have been shown by some of the plant scientists. (Fabio Minervini, et al. [100]) assessed the dynamics of lactic acid bacteria and other Firmicutes associated with durum wheat organs and processed products. 16S rRNA gene-based high-throughput sequencing showed Lactobacillus, Streptococcus, Enterococcus, and Lactococcus as the main epiphytic and endophytic genera among lactic acid bacteria. Bacillus, Exiguobacterium, Paenibacillus, and Staphylococcus completed the picture of the core genus microbiome. The relative abundance of each lactic acid bacterium genus was affected by cultivars, phenological stages, other Firmicutes genera, environmental temperature, and water activity (aw) of plant organs. Lactobacilli, showing the highest sensitivity to aw, markedly decrease during milk development (Odisseo) and physiological maturity (Saragolla). At these stages, Lactobacillus was mainly replaced by Streptococcus, Lactococcus, and Enterococcus. However, a key sourdough species, Lactobacillus plantarum, was associated with plant organs during the life cycle of Odisseo and Saragolla wheat. The composition of the sourdough microbiota and the overall quality of leavened baked goods are also determined throughout the phenological stages of wheat cultivation, with variations depending on environmental and agronomic factor (Li Zhou, et al. [101]) showed that the grape polyphenols promoted the changes in the relevant gut microbial populations and shifted the profiles of SCFAs. Fermentation of grape polyphenols resulted in a significant increase in the numbers of Bifidobacterium spp. and Lactobacillus–Enterococcus group and inhibition in the growth of the Clostridium histolyticum group and the Bacteroides–Prevotella group, with no significant effect on the population of total bacteria. The findings suggest that grape polyphenols have potential prebiotic effects on modulating the gut microbiota composition and generating SCFAs that contribute to the improvements of host health. (M Abror, et al. [102]) concluded that leri water and lactobacillus bacteria have an effect on the growth and production of mustard greens.
Most plant-associated strains belong to Lactococcus lactis subsp. lactis, whereas Lactococcus lactis subsp. cremoris is typically found in dairy fermentations (Kelly, et al. [103,104]). Fermenting plant material is a second important ecosystem occupied by L. lactis, where it typically occurs as an early colonizer that is later replaced by species that are more tolerant of low pH values (Kelly, et al. [103,104]). Fermenting plant material comprises a broad array of highly variable niches with respect to chemical composition, for instance the availability of carbohydrates other than lactose as growth substrates. Moreover, protein concentrations are typically much lower than those observed in the dairy environment. As a result, strains isolated from fermenting plant material do not harvest amino acids through proteolysis but depend on amino acid biosynthesis and consequently exhibit fewer amino acid auxotrophies than do dairy isolates (Ayad, et al. [105]). Therefore, it can be anticipated that strains adapted to the plant ecological niche will exhibit large metabolic differences and their metabolic diversity will most certainly exceed that of dairy strains. Recently, it has been shown that strains isolated from a non-dairy environment exhibit flavor-forming activities that may be beneficial to dairy fermentation, as exemplified by the production of the key flavor fusel aldehydes as a result of a unique α -keto acid decarboxylase activity (Smit, et al. [106,107]). Moreover, it was shown that some non-dairy L. lactis strains produce the enzyme glutamate dehydrogenase, which converts glutamate to α -ketoglutarate (Tanous, et al. [108]). This compound, α -ketoglutarate, is the acceptor of the amino group in aminotransferase reactions, the first step in the production of flavor compounds from amino acid, and present at rate-limiting concentrations in cheese (Tanous, et al. [108]).
Lactobacillus: Lactobacillus is a genus of lactic acid bacteria described as a heterogeneous group of regular non spore forming gram positive rods and found in a great variety of habitats such as plants and gastrointestinal tracts (Amin, et al. [109]). The author isolated lactobacilli from plants to determine their inhibitory effect against some pathogens. Sixty lactobacilli isolates from fresh vegetables were enriched in Man-Rogosa-Sharpe medium (MRS) broth and isolated by growing on MRS agar medium, and were characterized by phenotypic characteristics and PCR technique at genus and species levels. The antimicrobial substance was extracted with ethyl acetate solvent and the antimicrobial activity against some pathogenic bacteria such as Escherichia coli, Salmonella typhi, Shigella dysenteriae, Bacillus anthracis and Staphylococcus aureus were investigated. The antimicrobial compound of fourteen plantarum and eight L. casei isolated from fresh vegetables showed a potent inhibitory activity against all tested human pathogenic bacteria. The inhibitory substance was distinct from bacteriocins, lactic and acetic acids which are produced by these bacteria. In conclusion, fresh vegetables may be used as a source of antimicrobial lactic acid bacteria. L. casei and L. plantarum as two probiotics can establish themselves in gut and urogenital tract and prevent the human body from adverse effects of pathogens.
Enterococcus: On plants, enterococci occur in a truly epiphytic relationship (Mundt, et al. [110]). These Enterococcus species typically associated with plants include the yellow-pigmented mundtii and E. casseliflavus (Martin and Mundt, 1972). The early studies on enterococci (‘faecal streptococci’) occurring on plants by (Mundt, et al. [110]) were performed before the genus Enterococcus was re-defined by Schleifer and Kilpper-Bälz (1984). Modern, taxonomic studies based on molecular, biological techniques for classification and species identification by (Thomas Muller, et al. [111]) validated this epiphytic relationship and enterococci occurring on plants were identified as E. faecium, E. faecalis, E. casseliflavus, E. mundtii and E. sulfureus. The majority of the isolates in the study of (Thomas Muller, et al. [111]), however, possessed a 16S rDNA genotype uncommon to Enterococcus species described at the time of the study. Enterococci also occur on fresh produce and possibly originate from the use of untreated irrigation water or manure slurry for crop production (Johnston and Jaykus, 2004). Interestingly, in this context Johnston and Jaykus (2004) isolated mainly E. faecalis and E. faecium strains, but also other Enterococcus spp. from fresh produce such as celery, cilantro, mustard greens, spinach, collards, parsley, dill, cabbage and cantaluope, and showed that many strains harboured antibiotic resistances. Similarly Ronconi, et al. (2002) isolated predominantly E. faecium and E. faecalis strains from lettuce and many strains were also antibiotic resistant.
(Thomas Muller, et al. [111]) isolated the species of Enterococcus viz. E. faecium, E. mundtii, E. casseliflavus, E. faecalis and E. sulfureus from the plant habitat. The majority of isolates differed distinctly in their restriction patterns from those of known species. They formed a group of a homogeneous 16S rDNA genotype (VI). The taxonomical investigations suggest that the isolates of the 16S rDNA genotype VI represent a new plant-associated Enterococcus species. (Franz, et al. [112]) reported the occurrence of Enterococci in a wide variety of environmental niches including soil, surface waters, waste waters, municipal water treatment plants, on plants, and in the gastrointestinal tract of warm blooded animals (including humans) and, as a result of association with plants and animals, in human foods (Franz, et al. 1999). (Lee, et al. [113]) demonstrated the use of Enterococcus faecium strain LKE12 to enhance plant growth in oriental melon. This bacterial strain was isolated from soil, identified as E. faecium by 16S rDNA sequencing and phylogenetic analysis. The plant growth-promoting ability of LKE12 bacterial culture was tested in a gibberellin (GA)-deficient rice dwarf mutant (waito-C) and a normal GA biosynthesis rice cultivar (Hwayongbyeo). E. faecium LKE12 significantly improved the length and biomass of rice shoots in both normal and dwarf cultivars through the secretion of an array of gibberellins (GA1, GA3, GA7, GA8, GA9, GA12, GA19, GA20, GA24, and GA53), as well as indole-3-acetic acid (IAA). Increases in shoot and root lengths, plant fresh weight, and chlorophyll content promoted by E. faecium LKE12 and its cell-free extract inoculated in oriental melon plants revealed a favorable interaction of E. faecium LKE12 with plants. Higher plant growth rates and nutrient contents of magnesium, calcium, sodium, iron, manganese, silicon, zinc, and nitrogen were found in cell-free extract-treated plants than in control plants. These results suggest that E. faecium LKE12 promotes plant growth by producing GAs and IAA; interestingly, the exogenous application of its cell-free culture extract can be a potential strategy to accelerate plant growth.
Sacchoromyces: (Thierry, et al. [114]) demonstrated that plants perform rhizophagy, a process in which live microbial cells are engulfed by root cells and digested to acquire the nutrients from the microbes. The phenomenon can be observed in both dicotyledonous and monocotyledonous plants. The rhizophagy is an evolutionarily conserved trait that predates the divergence of dicot and monocot species. To explore the potential relevance and practical application of rhizophagy, the brewers’ yeast (Saccharomyces cerevisiae), a waste product of the brewing industry, was used for its role as biofertilizer. The addition of live or dead yeast to fertilized soil substantially increased the nitrogen (N) and phosphorus (P) content of roots and shoots of tomato (Solanum lycopersicum) and young sugarcane plants. Yeast addition to soil also increased the root-to-shoot ratio in both species and induced species-specific morphological changes that included increased tillering in sugarcane and greater shoot biomass in tomato plants. These findings support the notion that brewers’ yeast is a cost-effective plant growth promoting microbe which is also used as probiotics.
Use of Gut Microbes in Processed Food Industry
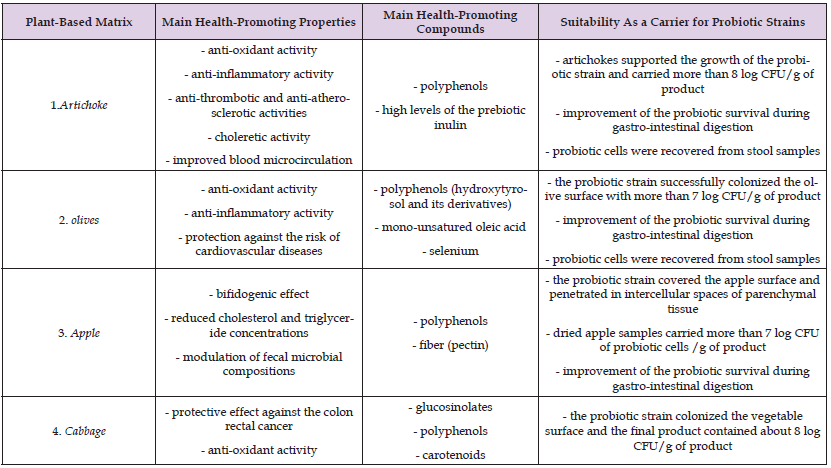
The performance of probiotic bacterial strains is influenced by the carrier food and its functional components which while buffering the probiotic through the gastro-intestinal tract, contribute to an efficient implantation of bacterial cells and regulate probiotic features (De Bellis, et al. [115]). Particularly, plant-based matrices are eligible substrate for hosting and delivering microbial populations because of their richness in nutrients, fibers, vitamins, minerals and dietary bioactive phytochemicals. The available data indicate that the intrinsic health-promoting properties of diverse plant-based matrices can be successfully exploited for developing effective association with probiotics. The health-promoting properties of solid plant-based matrices (particularly artichokes, table olives, apple and cabbage) and their association with probiotic bacteria are indicative of the role of the food matrix in sustaining probiotic cells during product processing, digestive process, gut implantation, and finally in exerting beneficial effects.
It should be considered that, since probiotics only transiently colonize the intestinal tract, large populations need to be daily ingested to provide health benefits (Hill, et al. [90]). In fact, probiotic survival during gastro-intestinal (GI) digestion and gut colonization suitability are strain-related abilities, and the efficacy of probiotic bacteria is influenced by the carrier food and its components, which while protecting the probiotic through the GI tract, contribute to an efficient implantation of bacterial cells and regulate probiotic attributes (Flach, et al. 2018, Ranadheera, et al. 2010). In this regard, vegetable matrices are eligible for hosting and delivering microbial populations and particularly probiotic strains which are also able to increase their intrinsic health-promoting and functional properties. In fact, the functional attributes of plant-based matrices, their structure and their suitability to fermentation make them appropriate for carrying probiotic strains that would take advantages from the characteristics of plant-based matrices and, by exploiting prebiotic and bioactive molecules, take benefit for their survival during product processing and shelf life as well as in the digestive process and gut colonization. The functional properties of plant-based matrices depend on their richness in nutrients, fibers, vitamins, minerals and dietary bioactive phytochemicals and some of those diverse components have also an important role in the interactions with gut microorganisms (Flach, van der Waal, van den Nieuwboer, Claassen, & Larsen, 2018). In particular, the fiber content (dietary fibers) is involved, directly as well as for its effect on the gut microbiota (Holscher, 2017, Simpson and Campbell, 2015), in a number of recognized health-promoting effects of plant-based foods. Phenolic compounds of the vegetable matrices have been also associated to plant health-promoting activities; moreover, their potential prebiotic activity as well as of their process-derived bioactive molecules have been recently recognized (Alves-Santos et al., 2020, Debelo et al., 2020). The most relevant plant-based matrices suitable as vectors for delivering Gut microbe /probiotics are described in (Tables 5 & 6).
Note: B.: Bifidobacterium; E.: Escherichia; L.: Lactobacillus; Lc.: Leuconostoc; Ls.: Listeria; S.: Saccharomyces.
Table 6: Main health-promoting properties and compounds of plant-based matrices suitable as carriers for probiotic strains.
Health Promoting Effects of the Association of Gut Microbe/Probiotic Bacteria with Some Plant Based Matrices:
1. Improved probiotic cell survival during gastro-intestinal digestion (e.g. artichoke, table olive, apple.
2. Improved gut colonization (e.g. artichoke, table olive)
3. Gut microbiota modulation (e.g. artichoke, table olive, apple)
4. Stimulation of resident microflora to produce SCFA (e.g. artichoke, table olive, apple)
5. Improved immune-modulatory activity (e.g. artichoke)
6. Relieving in constipation symptoms (e.g. artichoke)
7. Antioxidant activities (e.g. artichoke, table olive, apple, cabbage)
Concept of Human Gut Microbe Assisted Agricultural Food Production
It is evident that some of the gut microbes are associated with the crop plants in the agricultural production system with some unknown and known roles (Mundt, et al. [110,111,103,109,114,113,100,102]). These gut microbes are indispensable part of the human health and their population and equilibrium in the gut plays an important role in various metabolic, hormonal, and neurological functions. Although, these gut microbes are available in the form of probiotics, prebiotics are required for their proper multiplication and maintenance in the GI tract. Therefore, plant based matrix probiotic, as processed food, is available for this purpose (Betoret, et al. [116-118]). However, fresh crop produce particularly fruits and vegetables can be a source of these gut microbes, if these are used in the crop production system [119-221]. There is a need to explore their use in the form of probiotic sprays, on the salad vegetable and fruits, which are consumed raw for health benefits. At present, we do not know, whether these probiotic will survive as epiphytic or both as epiphytic and endophyte in the agriculture produce. Neither, all the available probiotics are studied for their plant growth promoting activities except the species of lactic acid bacteria. Therefore, there is an urgent need to explore their use in the agricultural production system and if we could produce the gut microbe lased fruits and vegetable, these can be a source of better nutrition to mankind and a remedy to solve many health issues which are govern by gut microbes.


