Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Congyue Zhang1,2, Chaoqun Zhang1,2, Huifang Zhou2, Zhankai Hu1 and Dianxing Sun2*
Received: December 22, 2022; Published: January 13, 2023
*Corresponding author: Dianxing Sun, The 980th Hospital of Chinese People’s Liberation Army Joint Logistics Support Force, Shijiazhuang, China
DOI: 10.26717/BJSTR.2023.48.007604
Background: Hepatocellular carcinoma (HCC) is a prevalent malignancy worldwide, and ferroptosis is an iron-dependent cell death process. In addition, the aberrant expression of long noncoding RNAs (lncRNAs) that contribute to the development and progression of HCC has garnered increased interest. Materials and Methods: We collected lncRNA expression profiles associated with ferroptosis and clinicopathological information from The Cancer Genome Atlas (TCGA) and FerrDb databases. The relationship between ferroptosis-related lncRNAs (FRlncRNAs) and HCC patients’ survival is determined by co-expression analysis of overall survival (OS). Using Cox regression analysis and the LASSO algorithm, a prognostic lncRNA model of 22 differentially expressed lncRNAs was developed. Results: high-risk lncRNA profile was associated with a poor prognosis in HCC, as determined by a Kaplan-Meier analysis. In predicting the prognosis of HCC, our risk assessment model outperformed conventional clinical data. GSEA uncovered immune and tumor-related pathways in high- and low-risk individuals. In addition, TCGA revealed that T cell functions, such as B cells, cytolytic macrophages, MHC-class-I, mast cells, neutrophils, NK cells, helper T cells, Type-I-IFN, and Type-II-IFN, differed significantly between high- and lowrisk groups. Immune checkpoints were also differentially expressed between the risk groups, including TNFSF18, IDO2, CD276, NRP1, and TNFSF4. Conclusion: Based on lncRNAs associated with ferroptosis, our findings provide a robust prognostic and immune response prediction model for HCC patients.
Keywords: Hepatocellular Carcinoma; Lncrna; Ferroptosis; Gene; Immune
Abbreviations: FRlncRNAs: Ferroptosis-Related lncRNAs; HCC: Hepatocellular Carcinoma; lncRNAs: Long Non-Coding RNAs; TCGA: The Cancer Genome Atlas; OS: Overall Survival; ROS: Reactive Oxygen Species; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; BP: Biological Processes; MF: Molecular Function; CC: Cellular Components; DEGs: Differentially Expressed Genes; GSEA: Gene Set Enrichment Analysis; FDR: False Discovery Rate; DCA: Decision Curve Analysis; ROC: Operating Characteristic Curve; ssGSEA: Single-Sample Gene Set Enrichment Analysis; HR: Hazard Ratio; CI: Confidence Interval.
HCC is the most prevalent pathological subtype of primary liver cancer, the fifth most prevalent malignancy globally, and the third leading cause of cancer-related mortality [1]. Multiple factors, including the chronic hepatitis virus, heavy alcohol consumption, and chronic hepatitis due to nonalcoholic fatty cirrhosis, contribute to the development of HCC [2]. Surgical resection or liver transplantation can effectively control cancer progression and prolong survival in patients with HCC at an early stage. Nonetheless, greater than two-thirds of patients will experience recurrence [3], and these patients are frequently diagnosed at an intermediate or advanced stage when recurrence occurs. Furthermore, most patients have fewer surgical options due to age or physical condition. Therefore, the effectiveness of interventional therapy, radiotherapy, targeted therapy, and local ablation is limited, and the prognosis remains dismal [4]. Consequently, it is essential to investigate the potential molecular mechanisms and cellular signaling pathways in the pathogenesis of HCC, seek early diagnosis and treatment, study the expressed genes with prognostic value, and develop a model with predictive characteristics.
In recent years, ferroptosis research has increased exponentially. It is a programmed cell death that is iron-oxidatively reactive oxygen species (ROS)-dependent and lipid peroxidation-mediated [5]. The significance of ferroptosis in regulating metabolism and redox biology has been demonstrated, influencing the pathogenesis and treatment of cancers such as prostate cancer, gastric cancer, and HCC. Recently, ferroptosis induction has emerged as a promising treatment for cancers resistant to conventional treatment. Zhang, et al. [6,7] discovered that the tumor suppressor BAP1 inhibited cystine uptake by inhibiting SLC7A11 expression, resulting in increased lipid peroxidation and ferroptosis Sun, et al. [8]. discovered that p62 expression inhibited nuclear factor degradation of erythroid 2-related factor (NRF2) and increased NRF2 nuclear accumulation via the misfiring of kelch-like ECH-associated protein 1. Liu, et al. [9]. reported the correlation between ferroptosis and the immune system and concluded that this prognostic factor could be used to screen HCC patients for immunotherapy and targeted therapies [10]. Long noncoding RNAs (LncRNAs) are noncoding transcripts longer than 200 nucleotides that can modulate the expression of numerous genes associated with cancer. Recent research by Sun et al. revealed that high levels of lncRNA GA-binding protein subunit (GABPB1) antisense RNA 1 in HCC cells enhanced erastin-induced ferroptosis by inhibiting GABPB1 translation and peroxidase-5, resulting in inhibition of cellular antioxidant capacity and cell survival [11]. Zhang et al. analyzed the relationship between ferroptosis and tumor mutations in HCC to develop a ferroptosisrelated gene model that may bridge the gap between ferroptosis and nuclear tumor mutations, thereby allowing for individualized treatment of HCC patients [12]. Using the TCGA database, this study constructed a model of prognosis-related lncRNAs. The role of ferroptosis-related lncRNAs, N6-methyladenosine (m6A) mRNA, and immune response in determining the prognosis of HCC has been investigated [13].
Data Sources and Clinical Information
From the TCGA database, RNA-sequence data and associated clinical data were extracted for 424 samples, including 374 HCC tissues and 50 normal liver tissues. An overview of the relevant clinical aspects of HCC patients can be found in Supplementary (Supplementary Table 1)., with 377 clinical data available for further analysis (Table 1). The collected clinical data for patients with HCC included their gender, age, grade, TNM stage, survival status, and survival time.
Identification of FRlncRNAs and Enrichment Analysis
The corresponding ferroptosis-related genes were downloaded from FerrDb [14], a web-based consortium offering an exhaustive and current database of ferroptosis markers, regulatory molecules, and associated diseases. We ultimately identified 382 ferroptosisrelated genes (Supplementary Table 2), of which Diver comprises 150, Suppressor comprises 109, and Marker comprises 123. Pearson’s test was applied to these genes to determine the relationship between FRlncRNAs and HCC. The association was deemed significant if the correlation coefficient |R2| > 0.5 and p < 0.001. First, the functions of up- and down-regulated ferroptosisrelated differentially expressed genes (DEGs) were investigated. Then, we analyzed the biological pathways associated with DEGs using the Kyoto Encyclopedia of Genes and Gene Expression (KEGG) data. According to gene ontology (GO), the functions of biological processes (BP), molecular functions (MF), and cellular components (CC) regulated by differentially expressed FRlncRNAs were investigated further. Using the GO and KEGG pathways, the «cluster Profiler» R package [15] assessed the biological roles of the prognostic candidates.
Construction and Validation of a Model of FRlncRNAs
Utilizing Lasso-penalized Cox regression analysis, FRlncRNAs with prognostic value were screened. In the final cohort for analysis, 376 lncRNA-seq samples remained after excluding those with unknown survival times (n = 1) based on previous clinical data. A genetic model containing biomarkers helpful in predicting prognosis was identified using the «glmnet» package in R. A risk score was calculated for each sample in all data sets based on this model, according to the risk score = alncRNA1 × lncRNA1 expression + blncRNA2 × lncRNA2 expression +⋯ + nlncRNAn × lncRNAn expression. To evaluate the predictive ability of the model for prognostic risk, we analyzed characteristic curves (ROC) for 10-year survival using the «timeROC» function in R. Based on median scores, the RNA was split into low- and high-risk groups. Using Kaplan-Meier analysis, the prognostic significance of this model for HCC was explored. Finally, univariate and multivariate Cox regression analyses were conducted to determine whether the model had good predictive power independent of other clinicopathological characteristics.
Predictive Nomogram
The R package «regplot» was utilized to compile clinical data and develop a nomogram integrating prognostic features to predict the 1-, 3-, and 5-year OS of HCC patients.
Function Enrichment Analysis
Based on gene set enrichment analysis, potential functional pathways involved in the ferroptosis lncRNA model were identified (GSEA). GSEA was performed on the KEGG dataset c2.cp.kegg. v7.4.symbols.gmt in java GSEA4.1.0 using the optimal cut-off value to divide TCGA data and identify enrichment pathways between the high-risk and low-risk groups. Statistical significance was set at p < 0.05, and false discovery rate (FDR) q < 0.25 was deemed statistically significant.
Immunity Infiltrates Analysis and Gene Expression
In addition, the TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC [16-22] algorithms were compared to evaluate FRlncRNA models between high-risk and low-risk groups based on cellular components and cellular immune responses. Heatmaps depicted the differences in immune responses under various algorithms. Moreover, single-sample gene set enrichment analysis (ssGSEA) was employed to quantify tumorinfiltrating immune cell subpopulations between the two groups and evaluate their immune function.
Cell Culture and qRT-PCR
Hepatic stellate cells LX-2 and HCC cells HepG2, Huh7 and Hep3B were cultured in DMEM medium (Gibco, China) supplemented with 10% fetal bovine serum (Gibco, China) and 1% penicillinstreptomycin (Gibco, China). The cultures were placed in a sterile incubator at 37 °C and 5% CO2. The cells were obtained in the logarithmic growth phase for subsequent experiments. Following the manufacturer’s instructions, total RNA was extracted using RNAiso reagent (TaKaRa, 9108, China), and cDNA synthesis was reversed using the Prime Script RT kit (TaKaRa, RR047A, China). qPCR assays were performed using TB Green® Premix Ex Taq™ (Takara, RR420A, China). β-ACTIN was used as an endogenous reference. Relative quantification of lncRNAs was calculated using 2-ΔΔCT. The primers’ sequences are listed in (Supplementary Table 3).
Statistical Analysis
All statistical analyses were conducted using the R programming language and associated software packages. Using ROC and decision curve analysis (DCA), [23] evaluated the relationship between the model and its clinicopathological performance. The statistical significance level for each analysis was set at p < 0.05.
Enrichment Analysis of Ferroptosis-Related Genes
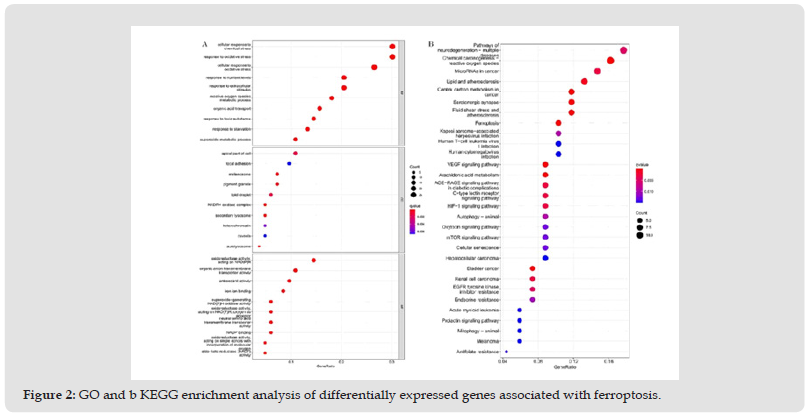
(Figure 1) depicts the study’s workflow. Supplementarily, we identified 84 DEGs associated with ferroptosis (13 downregulated and 71 upregulated; (Supplementary Table 4). GO enrichment analysis revealed that BP was involved in the cellular response to chemical and oxidative stress. The production of the apical portion of cells, focal adhesion, melanosomes, and pigment granules was primarily regulated by MF. NAD(P)H oxidoreductase activity, organic anion transmembrane transporter activity, and antioxidant activity were primarily upregulated in CC. KEGG pathway analysis revealed that the over-expressed genes were primarily involved in neurodegeneration and multiple disease pathways, chemical carcinogenesis and reactive oxygen species, microRNAs in cancer, lipid and atherosclerosis, central carbon metabolism in cancer, serotonergic synapse, fluid shear stress and atherosclerosis, and ferroptosis [24] (Figure 2a, 2b) and (Supplementary Table 5).
Figure 1 The study’s flow chart. TCGA: The Cancer Genome Atlas; ROC: operating haracteristic curve; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene ontology; GSEA: Gene set enrichment analysis; ssGSEA: single-sample gene set enrichment analysis.
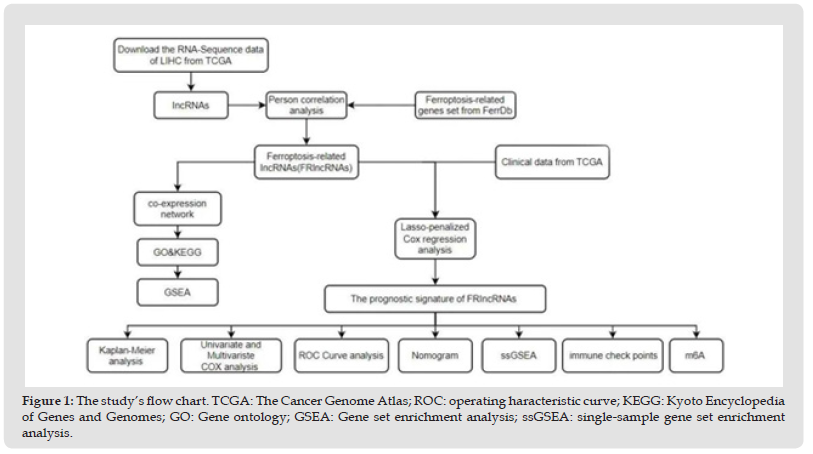
Figure 2 GO and b KEGG enrichment analysis of differentially expressed genes associated with ferroptosis.
Prognostic Characteristics and Prognostic Value of the Model
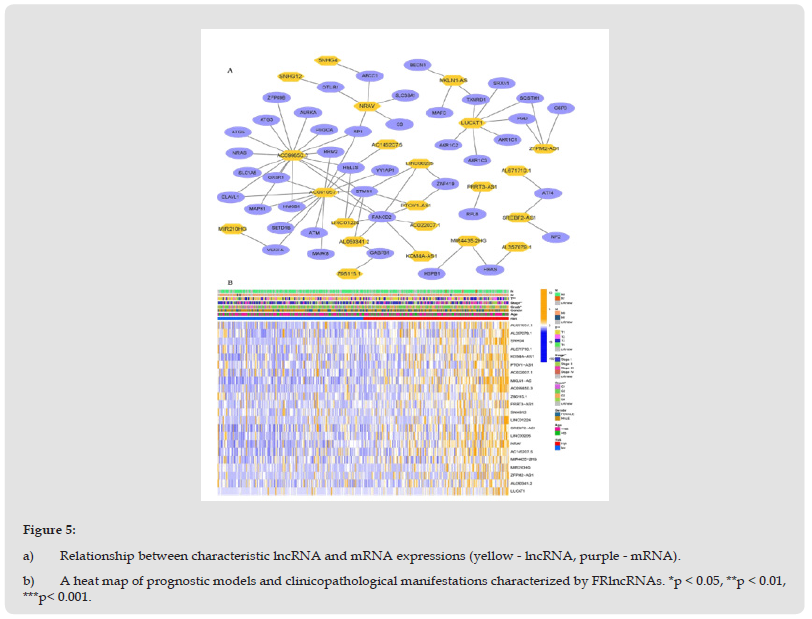
We identified 1072 lncRNAs associated with ferroptosis (Supplementary Table 5). Univariate COX analysis established 58 significant FRlncRNAs included in multivariate Cox analysis. Twenty-two DE-lncRNAs (AC091057.1, AL357079.1, SNHG4, AL671710.1, KDM4A-AS1, PTOV1-AS1, AC022007.1, MKLN1- AS, AC099850.3, Z95115.1, PRRT3-AS1 SNHG12, LINC01224, SREBF2-AS1, LINC00205, NRAV, AC145207.5, MIR4435-2HG, MIR210HG, ZFPM2-AS1, AL050341.2, LUCAT1) were determined as independent prognostic predictors of HCC (Supplementary Table 6). Consequently, we calculated the risk scores and constructed prognostic signatures for lncRNAs. Kaplan-Meier analysis confirmed that patients in the high-risk group with TCGA-LIHC had significantly shorter survival times at 1, 3, and 5 years compared to those in the low-risk group (p < 0.001, (Figure 3a)). In addition, the time-dependent ROC curve for survival prediction of the risk score model had an AUC of 0.811 at one year, which was more specific and sensitive in predicting the prognosis of HCC than conventional clinicopathological features (Figures 3b, 3e). In addition, timedependent ROC analysis demonstrated that the AUC predictive value of the novel model for the 1-, 3-, and 5-year survival rate was 0.811, 0.752, and 0.692, respectively (Figure 3d). The risk score and survival status of each HCC patient were represented using prognostic curves and scatter plots. Using patient risk survival status plots, we determined that patient risk score was negatively correlated with HCC patient survival and that most deaths occurred in the high-risk group (Figure 3c). Next, univariate Cox analysis revealed that lncRNA-based characteristics (HR: 1.106, 95% CI: 1.081-1.132) and tumor stage (HR: 1.680, 95% CI: 1.369-2.062) were independent prognostic factors in HCC patients (Figure 4a). Multivariate Cox analysis indicated that lncRNA characteristics (HR: 1.098, 95% CI: 1.072-1.125) and tumor stage (HR: 1.585, 95% CI: 1.282-1.958) were likewise independent prognostic risk factors for HCC patients (Figure 4b). (Figure 5a) depicts the relationship between lncRNAs and mRNAs. Additionally, we examined the association heatmap between prognostic characteristics and clinicopathological manifestations of FRlncRNAs (Figure 5b). In this combined nomogram, the risk score model was found to play the best role in these clinically significant variables and can therefore be used in the clinical prognostic assessment of HCC patients (Additional Figure 1). These studies demonstrate that this novel model of lncRNAs associated with ferroptosis is a reliable, independent prognostic factor for patients with HCC.
Figure 3 Based on FRlncRNAs signature: a) Kaplan-Meier curve results. b) AUC values of risk factors. c) risk survival status plot. d) AUC for predicting 1-, 3-, and 5-year survival of HCC. e) DCA of risk factors.
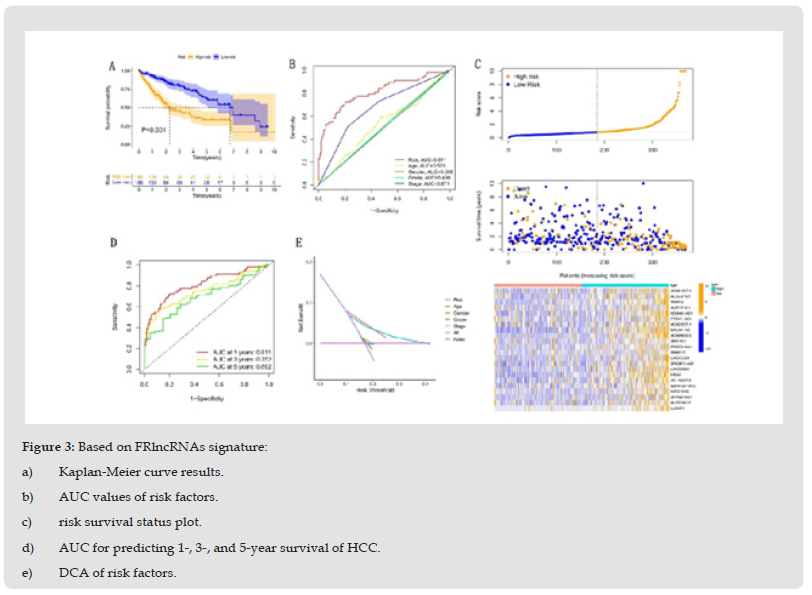
Figure 5 a) Relationship between characteristic lncRNA and mRNA expressions (yellow - lncRNA, purple - mRNA). b) A heat map of prognostic models and clinicopathological manifestations characterized by FRlncRNAs. *p < 0.05, **p < 0.01, ***p< 0.001.
GSEA Enrichment Analysis of Risk Scores
The majority of prognostic features of lncRNAs associated with ferroptosis regulate immune and tumor-related pathways, including ubiquitin-mediated proteolysis, homologous recombination, RIG- 1-like receptor signaling pathway, colorectal cancer, FC gamma R-mediated phagocytosis, TGF-β signaling pathway, notch signaling pathway, T cell receptor signaling pathway, regulation of autophagy, toll receptor signaling pathway, Aminoacyl tRNA biosynthesis, and RNA degradation (Supplementary Figure 2, 7).
Immunoassay and Gene Expression
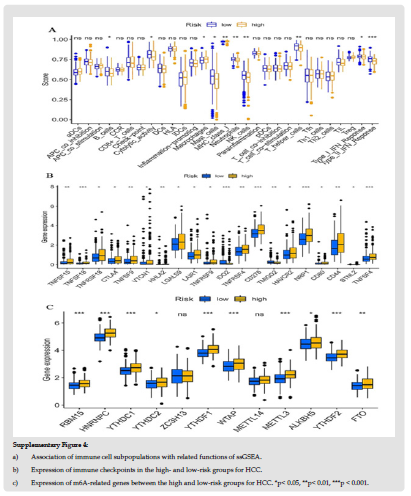
To determine whether the model is associated with tumor immunity, we compared the HCC risk score derived from the TIMER, CIBERSORT, CIBERSORT-ASS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms with an immunoreactive heatmap of tumorinfiltrating immune cells. The heatmap indicated that B cells, T cells, neutrophils granulocytes, macrophages, and medullary dendritic cells, including regulatory T cells, memory T cells, M0 macrophages, M2 macrophages, CD8+ T cells, lymphocytes, and CD4+ Th2 cells were significantly different between the high- and low-risk groups (Supplementary Figure 3). A correlation analysis of immune cell subpopulations and related functions in ssGSEA using the TCGALIHC database revealed that immune cells, including B cells, cytolytic macrophages, MHC-class-I, mast cells, neutrophils, NK cells, helper T cells, Type-I-IFN, and Type-II-IFN differed significantly between high- and low-risk groups (Supplementary Figure 4a). Considering the importance of immune checkpoint blockade based therapeutic strategies in HCC, we further investigated the differences in immune checkpoint expression between the high-and low-risk groups and found significant differences in the expressions of TNFSF18, IDO2, CD276, NRP1, and TNFSF4 between the two groups of patients (Supplementary Figure 4b). The comparison of m6A-related mRNA expressions between those two groups exhibited that RBM15, HNRNPC, YTHDC1, YTHDF1, WTAP, METTL3, ALKBH5, YTHDF2, and FTO were significant (Supplementary Figure 4c).
Expression of FRlncRNAs in HCC
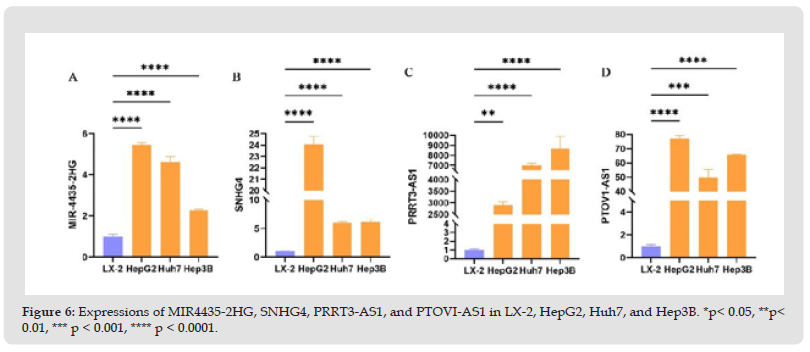
The expression of ferroptosis-related lncRNAs in hepatocellular carcinoma was validated by a series of bioinformatic analyses of 22 lncRNAs in patients with hepatocellular carcinoma and a predictive model constructed from prognosis-related differential ferroptosis genes. Based on this result, further experimental validation of these 22 genes was conducted. Before experimental validation, these 22 genes were independently researched in literature databases using the terms gene names and cancer. However, MIR4435-2HG, SNHG4, PRRT3-AS1, and PTOV1-AS1 were studied less in HCC. Consequently, in the subsequent experimental validation, these lncRNAs were logically chosen as gene targets for additional experimental proof of their relevance in HCC. HCC cell lines and hepatic stellate cell lines were used to validate the expression level of this lncRNA to investigate the expression of MIR4435-2HG, SNHG4, PRRT2-AS1, and PTOV1-AS1. The results of quantitative real-time PCR (qRTPCR) analysis revealed that these lncRNAs were «upregulated» in HCC cell lines relative to hepatic stellate cell lines (Figure 6). In addition, these results indicated that these lncRNAs might play a crucial role in HCC.
Figure 6 Expressions of MIR4435-2HG, SNHG4, PRRT3-AS1, and PTOVI-AS1 in LX-2, HepG2, Huh7, and Hep3B. *p< 0.05, **p< 0.01, *** p < 0.001, **** p < 0.0001.
In many cancers, such as gastric cancer [25] and head and neck squamous cell carcinoma, models of lncRNAs for predicting prognosis have been validated in previous studies. Ferroptosis can overcome the chemotherapy resistance of malignant cells and promote elimination of defective cells. Therefore, it may be a novel method for treating tumors. Based on the TCGA dataset, this study identified a novel model of prognostic lncRNAs associated with ferroptosis. The role of immune infiltrating cells in the tumor microenvironment and immune checkpoint inhibitors in the prognosis of HCC was subsequently investigated. Finally, this study identified potential biomarkers and therapeutic targets in the ferroptosis pathway. Our analysis revealed a total of 84 DEGs associated with ferroptosis. KEGG analysis revealed that these genes are primarily involved in neurodegeneration—multiple disease pathways, chemical carcinogenesis-reactive oxygen species, microRNAs in cancer, lipid and atherosclerosis, central carbon metabolism in cancer, serotonergic synapse, fluid shear stress and atherosclerosis, and ferroptosis. A recent study found that SREBP1/ SCD1-mediated lipid formation protects cancer cells from oxidative stress and ferroptosis when PI3K-AKT-mTOR signaling mutations are activated [26]. Fin56 is an inducer of type 3 ferroptosis, and Torin2 is a potent mTOR inhibitor used to activate autophagy, which has synergistic effects on the cytotoxicity of bladder cancer cells, as reported by Sun, et al. [27]. Recent research [28] discovered that ischemia-reperfusion (I/R)-induced upregulation of miR182-5p and miR378a-3p led to ferroptosis activation in kidney injury by downregulating GPX4 and SLC7A11. Metformin was found to induce ferroptosis in a mouse breast cancer xenograft model by upregulating miR324-3p [29]. In addition to isoglochidonine-induced ferroptosis, the miR122-5p/TP53/SLC7A11 pathway prevents brain hemorrhage-induced ferroptosis in neuronal cells [30]. In brief, our study identified 22 differentially expressed lncRNAs as independent HCC prognostic factors. KDM4A-AS1 protects AR and AR splice variants (AR/AR-Vs) from MDM2-mediated degradation of ubiquitin protectors, according to recent research. In addition, KDM4A-AS1 enhanced enzalutamide resistance in desmoplastic prostate cancer by inhibiting AR/AR-V degradation, and antisense oligonucleotide drugs targeting KDM4A-AS1 significantly reduced the growth of enzalutamide-resistant tumors [31]. A novel transcriptional gene, Yin Yang-1 (YY1), LINC00205, can accelerate HCC cell proliferation by sponging miR26a-5p to promote CDK6 expression [32]. LIN01224 absorbs miR193a-5p in gastric cancer and targets the upregulation of CDK8 to hasten the malignant transformation of gastric cancer [33]. LUCAT1 promoted TSCC cell proliferation, cell cycle, and migration by targeting miR375 [34]. Knockdown of LUCAT1 increased miR375 expression in tongue squamous cell carcinoma (TSCC) cells, and low miR374 expression was associated with a poor prognosis in TSCC. Overexpression of CACNA2D2 significantly inhibits non-small-cell lung cancer cell proliferation [35]. MIR210HG recruits DNA methyltransferase 1, which promotes methylation of the CACNA2D2 promoter region. In gastric cancer cells, LncRNA MIR44435-2HG binds to and inhibits the decapentaplegic protein (DSP), activating WNT/β-catenin signaling and triggering epithelial-mesenchymal transition [36]. Overexpression of the LncRNA muskelin1 antisense RNA (MKLN1- AS) increases the stability of Yes-associated transcriptional regulator 1 (YAP1) and promotes the proliferation, metastasis, and invasion of hepatocellular carcinoma cells [37]. LncRNA NRAV stimulates the production of the respiratory syncytial virus by sponging the miR-509-3p profile [38]. Silencing LncRNA PRRT3-AS1 activates peroxisome proliferator-activated receptor γ (PPARγ), inhibiting the mTOR signaling pathway to promote apoptosis and autophagy and inhibit prostate cancer cell proliferation [39]. By promoting LPS-induced inflammation in human lung fibroblasts and mouse lung tissue in vitro and in vivo, LncRNA SNHG4 suppresses METTL- 3-mediated transcriptional activator 2 (SATA2) mRNA at m6A levels [40]. Through regulation of the miR-15a/PDK4 axis [41], SNHG12 promotes colon carcinogenesis and progression. STAT1- inducible lncRNA ZFPM2 antisense RNA1 (ZFPM2-AS1) promotes colon cancer development by regulating the target of miR-653, the gene GOLM1, to reverse the inhibitory effect of miR-653 on the proliferation and metastasis of HCC cells [42]. However, few studies have examined the role of FRlncRNAs in HCC prognosis. The outcomes of our study may provide valuable insights for future cancer prevention efforts.
This study investigated ferroptosis biomarkers that aid in predicting the prognosis of HCC, which could serve as a reliable treatment guide for the disease. However, our model, primarily based on bioinformatics research, lacks experimental validation of these indicators. Therefore, additional validation employing distinct cohorts is required. Furthermore, given that our findings have not been validated using clinical samples, we cannot fully guarantee the dependability of our findings. In addition, the model’s established prognostic indicators require further validation [43].
In conclusion, we developed a model of FRlncRNAs for predicting prognosis and immune response in HCC patients, which was strongly correlated with a risk score, survival time, and clinical data on cancer. Consequently, the results suggest that our model of FRlncRNAs provides a personalized, predictive instrument for prognosis and immune response in HCC patients.
(Supplementary Tables 1-8), (Supplementary Figures 1-4)
Figure 7 Supplementary Figure 1: A Nomogram of FRlncRNAs for clinicopathological factors and prognosis.
Figure 9 Supplementary Figure 3: A heat map of immune responses in high- and low-risk groups based on TIMER, CIBERSORT, CIBERSORT-ASS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms. *p< 0.05, **p< 0.01, ***p<0.001.
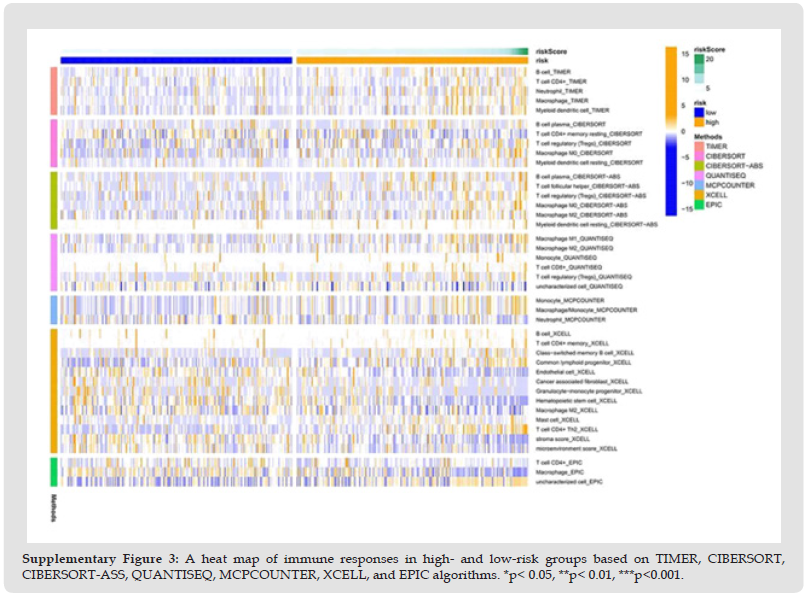
Figure 10 Supplementary Figure 4: a) Association of immune cell subpopulations with related functions of ssGSEA. b) Expression of immune checkpoints in the high- and low-risk groups for HCC. c) Expression of m6A-related genes between the high and low-risk groups for HCC. *p< 0.05, **p< 0.01, ***p < 0.001.
This manuscript was submitted as a pre-print with the link https://www.researchsquare.com/article/rs-1166940/v1.
We acknowledge the TCGA and FerrDb databases for providing their platforms and the contributors for uploading their valuable datasets.
This study was supported by grants from the Commission of China-Hebei Provincial Health Commission (No. 20211094).
Congyue Zhang conducted a literature review, gathered data, and designed and wrote the manuscript. Zhankai Hu aided in data collection. Huifang Zhou and Chaoqun Zhang reviewed the paper’s draft and suggested revisions. Finally, Sun Dianxing revised the manuscript critically. All authors have read and approved the final manuscript.
The raw data for this study were obtained from the TCGA database (https://portal.gdc.cancer.gov/), the specific sample is TCGA-LIHC, and the accession number is listed in the Supplementary file, which is a publicly accessible database.
Ethics approval and consent to participate: Both TCGA and FerrDb are public databases. Patients participating in the database have received ethical approval. Users can download relevant data for free and publish relevant articles for research purposes. Since our study is based on open-source data, there are no ethical or other conflicts of interest. We confirm that all procedures were executed following applicable rules and regulations. This study was conducted following the World Medical Association Declaration of Helsinki.
Not applicable.
The authors declare that they have no competing interests, and all authors attest to its accuracy.
associations. Database : the journal of biological databases and curation 2020: baaa021.


