Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Nuha Elagib Abdalla Elagib1, Nageeb Suliman Saeed2, Elamin Mohamed Ibrahim3 and Mohammed Ibrahim Saeed4*
Received: December 20, 2022; Published: January 13, 2023
*Corresponding author: Mohammed Ibrahim Saeed, Microbiology Department, Faculty of Medical Laboratory Science, The National Ribat University, Khartoum, 11111, Sudan
DOI: 10.26717/BJSTR.2023.48.007596
Campylobacter is a pathogen causing diarrhea and food-borne gastroenteritis worldwide in humans and animals and mainly causes childhood bloody diarrhea, anorexia, and abdominal pain.
Methods: This work focus on identifying Campylobacter, its antimicrobial susceptibility and relevant resistant genes in fecal specimen from infected children attending in Khartoum State hospitals using direct microscopy, isolation, identification, disc diffusion sensitivity test and resistance gene detection in PCR. 150 Fecal specimens were collected in Cary-Blair transport medium, and inoculated in Skrriow selective medium containing 5% defibrinated sheep blood, with selective antibiotic supplement; incubated for 48- to72 hrs at 42˚C in a microaerophilic condition in anaerobic jar using Gas-pack kits. The antibiotic susceptibility Kirby-Bauer assay was performed in Muller & Hinton with 5% blood agar against seven different antibiotics. While the antibiotic resistance genes for Ciprofloxacin (gyrA), Tetracycline tet (O), and Gentamicin (Gen R) were determined by PCR.
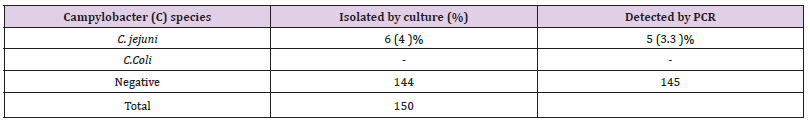
Results: All Campylobacter isolates were jejuni out of 150 cultured stool specimens from children revealed were five isolates (3.3%). Six C. jejuni were detected by PCR (4%). Campylobacter Coli was not detected. The antibiotic susceptibility pattern revealed that all C. jejuni isolates were susceptible to Erythromycin Nalidixic acid, Tetracycline, Amoxacillin and Ciprofloxacin. But showed intermediate resistance to Gentamicin that was confirmed by the detection of Gentamicin-resistant genes Gen (R) in PCR. While; tetracycline (tet O), and Ciprofloxacin (gyrA) resistance genes were negative.
Conclusion: Campylobacter jejuni was the only identified species circulating and causing diarrhea among children 2 - 5 years. The isolates harbor Gentamicin resistant genes, but luckily they were susceptible to Erythromycin, Nalidixic acid, Tetracycline, Amoxacillin, and Ciprofloxacin. In addition, the molecular assay was more sensitive in pathogen detection, identification, and determination of drug resistance genes in Campylobacter jejuni compared to culture method.
Keywords: Antimicrobial; Gentamicin-Resistant; Genes; Campylobacter-Jejuni; Children; Khartoum
The genus Campylobacter comprises a group of Gram-negative, curved rod-shaped bacteria. Most species prefer a micro-aerobic atmosphere containing 3-10 % oxygen for growth, [1]. In 1972, Campylobacter jejuni was first isolated from human diarrheal stool specimens by a clinical microbiologist in Belgium [2]. They primarily colonize the gastrointestinal tracts of a wide variety of animal host species, but C. jejuni and C. coli are the main-established causes of diarrhea in humans [3]. In humans, C. jejuni is the main cause of Campylo-bacteriosis, an enteric illness that can be transmitted to humans through the consumption of undercooked meat, especially poultry, contaminated water and milk, and contact with farm animals such as poultry and livestock [4]. In immunocompromised individuals, the infection is usually more severe [5]. Patients with C. jejuni or C. coli infection experience acute watery or bloody diarrhea, fever, weight loss, and cramps that last, on average, 6 days. And those who ingest the organism from contaminated food or water are at risk of acquiring the disease [1]. The majority of reported clinical cases were due to Campylobacter jejuni (90% to 95 %) and Campylobacter coli (5% to 10%) [6]. In industrialized countries, Campylobacter most commonly affects adults, mainly because of traveling. While, in tropical developing countries, it is more prevalent among young children between months to 5 years of age [7].
New Zealand had one of the highest human campylo-bacteriosis cases among all developed countries, [8]. Other reports, an increased incidence of human Campylo-bacteriosis cases in Denmark, it was a three-fold increase with C. jejuni or C. Coli can occur in patients of all ages, 1999, [9]. In Canada, 2004, Australia in 2005, and Germany, 2009 [10,11]. In 2007, C. jejuni was the common cause (15.7%) of acute gastroenteritis in infants and preschoolers in Mexico and Guatemala [12]. An investigation in Yangzhou, China, between July 2005 and December 2006 showed that 4.84% of 3,061 patients with diarrhea were PCR positive for C. jejuni, [13]. Among travelers; Campylobacter infection isolation rates were 39% and the frequency of C. jejuni infection was much higher in Japan than in the U.S. While; the incidence in England was quite high, 40,000 cases, in 2008, [1,12]. In Africa; the reported prevalence of Campylobacter in children under the age of five years; in South Africa, it was 17.3% in 202 patients, 2013. In Liberia, Campylobacter isolation rate of 44.9% in children from a crowded urban compared with 28% in cleaner rural area, [2]. In 2012 at a hospital in Kisii, Kenya, culture positive for Campylobacter species, was significantly higher [14]. Among Tanzanian children, the reported detection of C. jejuni was 34.8% of gastroenteritis cases and non-jejuni/coli Campylobacter species was 47.8% [15].
Campylobacter infection reported in Sudan as 2% in 437 stool samples of diarrheal children, 2013, [16]. In a previously reported study, out of two hundred 200 Sudanese children, 4.8 % were found having C. jejuni infection [17]. In another study of Campylobacter jejuni were 3.3% multiplex PCR assay from goats in 2015. [18]. For isolation of C. jejuni and C. Coli from feces required the Selective medium a blood agar in the Laboratory; routine diagnosis made by direct microscopy examination of Gram stained fecal sample, or by isolation of the organism in Skirrow’s selective medium, which contains Campylobacter agar base, 5% horse blood, Vancomycin, trimethoprim, and polymyxin B [19]. Cephalosporin to discourage the growth of Proteus species in addition to antifungal agent, The growth time is usually two to four days in a microaerobic atmosphere at 42±1 ºC [20]. Oxidase-positive and detection of l-alanine aminopeptidase activity are employed to differentiate between Campylobacter, Helicobacter and Arcobacter species and other Gram-negative bacteria [21]. C. jejuni hydrolyzes hippurate while hippurate while C. coli does not. While the biotype 2 of C. jejuni produces hydrogen sulfide in iron-containing medium, whereas C jejuni biotype 1 does not [22]. Most patients infected with Campylobacter spp. recover without any specific treatment other than replacing lost fluids and electrolytes. Most Campylobacter species are resistant to cefalothin; to which most other stool flora are susceptible [23]. The antimicrobial agents prescribed for Campylobacter infection are Erythromycin, for children, and fluoroquinolone, such as Ciprofloxacin, for adults. Campylobacter infections, antibiotic treatment in cases of severe illness or persistent enteritis. Erythromycin or fluoroquinolones are usually the antibiotics of choice. The Antibiotics of choice generally are macrolides, with tetracycline and (Fluoro) quinolones, are reserved for more severe cases. [24]. The growing increase in resistance to (fluoro) quinolones, tetracycline and erythromycin of C. coli and C. jejuni strains, might compromise the efficacy of this treatment.
Gentamicin remained the only alternative to fluoroquinolones and macrolides, for systemic infections caused by Campylobacter spp. [25]. Antibiotics have been in discriminately used in animal production for decades in order to control, prevent and treat infections, and enhance animal growth. A higher prevalence of multiresistant Campylobacter strains has been reported for animal and meat isolates than for human isolates [25]. In 1995, the incidence of fluoroquinolone resistance in Campylobacter isolates from Thailand was reported as 84%. And in 1997–1998, the incidence of fluoroquinolone resistance in Spain was reported as 72%. Incidence of resistance to the fluoroquinolones has also increased in the United States, United Kingdom, and the Netherlands, where the proportion of Campylobacter isolates resistant to the fluoroquinolones was reported as 10%, 18%, and 29%, respectively [22]. Fluoroquinolone-resistant C. jejuni were recognized during the late 1980s in Europe, where researchers suggested that such resistance was due, in part, to the acquisition of fluoroquinolone-resistant strains from animal sources [26]. In northern India, 2.2% of Campylobacter strains had multidrug resistances between 1989 and 1993. In the year 2005, the same group reported that MDR had increased to 30.6% among their C. jejuni and C. Coli strains collected during the year 2002 [27].
Rationale
Campylobacter infection in children is an important health problem, though C.jejuni are a common cause of gastroenteritis in Sudan. And the prevalence was not determined yet due to lack of proper surveillance system, and microbiology laboratories services would assistant in proper surveillance systems of Campylobacter infections, to determine the antibiotic resistance of pathogenic Campylobacter in Sudan.
The study aims were to detect and identify Campylobacter species infecting children attending hospitals in Khartoum State by conventional methods. And to determine their antibiotic susceptibility patterns and antibiotics-resistant genes using PCR based technique. Participants were children in the age range (1 to 12) years, presenting with diarrhea and acute gastrointestinal infection. The specimen was collected in a wide mouth container for isolation and identification of Campylobacter from stool on selective medium. Swabs were immediately inoculated into transport and later subcultures on Campylobacter selective Skirrow agar. Incubated under microaerophilic conditions using a commercial gas generating kits (CAMP gas generation) and incubated at 42°C temperatures. The suspected colonies, showing typical Campylobacter morphology and positive oxidase, were microscopically examined after Gram-staining and primarily identified through the sodium hippurate hydrolysis test [19]. Antibiotic susceptibility assay was performed using Kirby-Bauer disk diffusion test. Antibiotic panel used includes Erythromycin, Azithromycin, Gentamicin, Tetracycline, Ciprofloxacin, Nalidixic acid and Amoxacillin /clavulanic acid. The susceptibility of the isolates to each antibiotic was determined according to the latest guidelines published by the Clinical and Laboratory Standards Institute. (CLSI), [24]. DNA extraction was performed by boiling methods and PCR were performed from pure culture of each isolate and further identified by a PCR reaction used to amplify C. jejuni and C. Coli.
Specimens Collection
Stool samples were collected from children after informed consent from hospitals records or parents in a sterile disinfectant-free, screw-capped, wide necked containers and placed immediately into Cary-Blair transport media and submitted to the Microbiology Laboratory, at the National Health laboratory.
Macroscopic Examination
All specimens in plastic containers were examined macroscopically for its consistency, color, and the presence of mucus and blood.
Microscopic Examination
A wet mount of each specimen was prepared in normal saline on a slide with cover slip and examined under the microscope.
Culture
Swab from fecal specimen inoculated in Cary-Blair transport medium subcultures on skirrow medium plates and into brain Heart infusion broth from Oxoid, United Kingdom. Then incubation plates were incubated at 42ºC for 48 hours under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) achieved in anaerobic jar using microaerophilic Campylobacter gas generating kits. Identifications: the plates were examined for the morphology of suspected colonies with gray in ceramic, moist, slightly raised, drops like and with or without metallic sheen colonies were subjected to further identification by microscopic examination and Gram stain.
Biochemical Tests
The oxidase test was applied using reagent 1% (wt/ vol) solution of N, N, N’, N’–tetramethyl-p-phenylenediamine dihydrochloride, and development of purple colonies' reaction within 10 seconds considered positive. Catalase test performed using 3% hydrogen peroxide solution (H2O2) to give a positive reaction of air bubble. Motility test: from pure colonies were stabbed and incubated in tubes containing semisolid medium agar and the medium then incubated for 48/hrs. The positive motility test indicated by the spread of the growth to the surface and spread into the bottom. Hippurate hydrolysis assay: Used for identification and differentiation between the two species, C. jejuni and C. coli. The C. jejuni has ability to hydrolyze hippurate, but not C. Coli. A loop full of pure culture suspended in 2ml of distill water was added to 0.5ml of 1% sodium hippurate solution and incubated at 37°C for 10-14 minutes, Then 0.2ml of nine Hydrin solution was added on the top of each test tube above the hippurate solution. After incubated for 2 hrs at room temperature, then development of deep purple color indicating the Campylobacter due to hydrolysis of the hippurate considered as positive test for C. jejuni.
Antimicrobial Susceptibility Testing
Kirby-Bauer disk diffusion antibiotic susceptibility assay performed by using Mueller - Hinton agar with the addition of 5% blood agar medium was determined according to guidelines published by the Clinical Laboratory Standards Institute (CLSI). And 0.5 McFarland turbidity standard was prepared to control inoculum size during antimicrobial susceptibility testing. Seven antibiotic were examined including Erythromycin (15μg), Azithromycin (15μg), Gentamicin (10μg), Tetracycline (5μg), Ciprofloxacin (5μg), Nalidixic acid (30μg), and Amoxicillin/Clavulanic acid (30μg). The plate was incubated at 42 °C for 16–18 hours and the diameter of the zones of inhibition was measured.
DNA was extracted from the bacterial cell pellet by boiling methods, and DNA concentrations were determined by measuring the optical density at 260 nm by the nano-drop Analytica machine.
Boiling Methods
1 ml aliquots enrichment broth was centrifuged at 12,000 ramps for 2 minutes, then the pellets were suspended in 50 μl TE buffer, and heated at 95°C for 10 minutes on Heat dry block machine and centrifugated at 12000 rmps for 2 mins, the supernatant was transferred into a new sterile tube for PCR, [18].
DNA Extraction from Stool Sample
Stool suspended in a 2 ml brain Heart infusion broth in Eppendorf tube and centrifugated, 100 μl of 1x (Phosphate-Buffer-Saline) was added and the suspension thoroughly vortexes, heated for 5 min at 95°C and stool particles were pelleted by centrifugation. The supernatant was added to a new micro-centrifuge tube and 1x PBS was added and samples were vortexes then incubated for 1 minute at room temperature. The samples were then spun down to pellet the DNA. Finally, the spin the samples were placed in a new tube and 200 μL of TE Buffer added, incubated for 1 minute at room temperature and then centrifuged at full speed for 1 minute to elute DNA. The eluted DNA was stored at -20°C until PCR, [18].
PCR polymerase Chain reaction: The PCR assay was used for detecting C. jejuni and C. Coli. A primer pair was designed for co-identification of the two species (C. jejuni and C. Coli).
The genes were selected for identification of the genus Campylobacter species:
The C. jejuni primer sequence: -(5’-3’) Forward -GGAGAGGGTTTGGGTGGT and (3’-5’) Reverse -AGCTAGCCTCGCATAATAACTTG. And C. Coli: primer sequence - (5’-3’) Forwards -TTTTTAGCAAAGATTCTGAT and (3’-5”) Reverse primer sequence -CAAAGCATCATAAACTGCAA [28].
The gyrA resistance gene in Campylobacter for Ciprofloxacin Gene (R) for Gentamicin was tested using; Forwards primer sequence (5”-3’)) TTTTTAGCAAAGATTCTGAT and Reverse primer sequence (3′-’5”) CAAAGCATCATAAACTGCAA. Primers of the Gentamicin resistance gene used were; Forward primer F ((5′ -3′) AGTTGACCCAGGGCTGTCGC and Reverse primer R (3′-5′) GTGTGCTCTGGTCCACAGC. The Detection of Resistance Genes for Tetracycline(tetO) using forwards primer sequence F (5’--3’) AACTTAGGGGCATTCTGGCTCAC & reverse primer sequence R (3′-5) TCCCACTGTTCCATATCGTCA [28-30]. Agarose Gel Electrophoresis: the Campylobacter polymerase chain reaction (PCR) products were separated and checked for DNA size in 1.5 % agarose gel electrophoresis containing 0.1 μg/ml ethidium bromide [26]. Statistical analysis Data were entered using Excel and Access software’s ;s and analyzed using SPSS version 20 (IBM SPSS Statistics 20) to verify data and test to check the variances' equality of numeric data. The differences were considered significant when the P value was less than 0.05
In developing countries, where the isolation of Campylobacter and the rate of infection range from 5 to 20%; due to variations on sort feeding and nutrition system. Young children remain most susceptible, both in the developing and the developed world. Only six (6) isolates strains were identified as C. jejuni in PCR and The result showed that (4%) the rates of isolating Campylobacter jejuni in Khartoum state-Sudan this was less than previously reported by Oberhelman and Taylor in 2000 [11]. (Table -2 & Figure 1). Among study participants, their demographic data of Age group showed that more than 60% of children were sat age range from 3-5 yrs and the Gender show that 70% were males. The highest frequency of affected children were from Khartoum North Teaching Hospital. Bahri 60%, and Omdurman Teaching Hospital 20% (Figures 1 & 2).
Figure 1 Hippurate hydrolysis Test: Campylobacter jejuni: Formation of purple color indicative of the positive hippurate hydrolysis test. Wright: positive test in deep purple color, hydrolysis of the hippurate. Left site a colorless tube negative control. And in the middle was control positive test.

Campylobacter Isolation and Identification
The identified isolates showed the following laboratory test-based features; Five isolated strains were positive morphologically in culture skirrow selective medium; their colony was flat moist, translucent like water droplets. They have shown curved Gram-negative bacilli appearing in S -shape in Gram stain. They were motile, oxidase and catalase positive. And the five isolated strains shown hydrolysis of hippurate purple color, (Figure 1). Those outcomes indicated that out of 150 cultured fecal specimens, there were Five (5) (3.3%) identified as Campylobacter jejuni, (Table 1). While on PCR-based assay identified six positively detected C.jejuni strains out of 150 fecal samples, which represents a (4 %) table, figure. The Campylobacter Coli was not isolated from culture or detected in PCR, (Table 1).
Table 1: Percentage and number isolates of Campylobacter species from stool sample in Khartoum -state.
Antimicrobial Susceptibility Testing
The five isolates of Campylobacter jejuni were sensitive to Nalidixic, Azithromycin, Tetracycline, Erythromycine, Ciprofloxacine, and Amoxacillin acid, as shown by the increased zone diameter of inhibition compared to zone cut-off. The isolates were intermediately susceptible to Gentamicin and this resistance gene toward this antibiotic was confirmed by colony PCR method in AL isolates (Figure 2). While the Tetracycline and Ciprofloxacin resistance genes showed, were undetected, (Table 2 and Figure 3).
Figure 3 Antimicrobial Susceptibility Test for C. jejuni isolated from Khartoum state: The results demonstrated that (100%) of Campylobacter C.jejuni isolates were susceptible to Nalidixic Acid, ciprofloxacin, Azithromycin, Tetracycline, and Erythromycin. Intermediate resistant to Gentamicin (CN), observed in all C. jejuni isolates as shown in 2nd left.
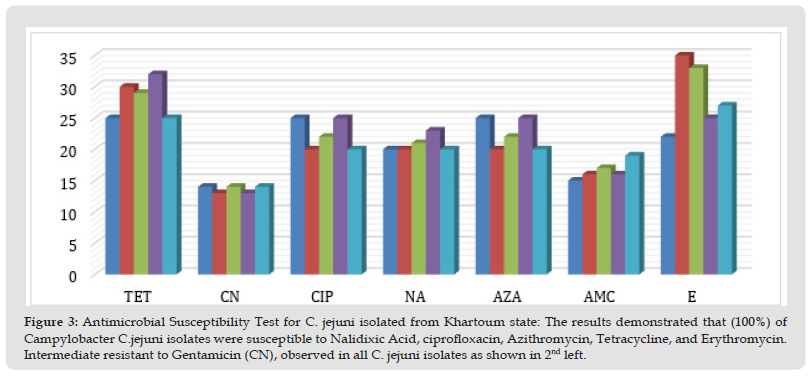
Note: Sensitive to: Nalidixic Acid, Erythromycin, Tetracycline, and Azithromycin, Intermediate to: Gentamycine & ciprofloxacin.
Campylobacter jejuni isolates were susceptible to a wide variety of antibiotics options, including: (Erythromycine), Amoxicillin, Azithromycin, tetracycline, ciprofloxacin. The molecular method has fastened lab results to three hours compared to 24 culture, and even it detects one non-viable and non-cultivable Campylobacter jejuni in one sample. In addition, it helps to check the type of drug resistance genes, to facilitate adequate treatment. The idetection rate of C. jejuni in Sudan was (4%) was agreed with Friedman, et al., finding in 2000 [31]. The prevalence of C. jejuni that isolated in other developing countries. C. Coli isolates were not detected in children in Khartoum – Sudan. The study finding was in agreed with Colles et al.a report in 2016, C. jejuni is responsible for the majority of infections in human clinical samples, but disagrees with the low incidence of C. Coli [32]. And agreed with Totten et al., in 2001 that the PCR amplification identified C jejuni isolates, and it confirmed the phenotypic results [33]. And can also provide an effective species level identification marker for this pathogen, especially when certain strains that may fail to hydrolysis hippurate and test negatively, which will be miss identified as C. Coli. The susceptibility results were in agreement with Ghunaim et al., 2015 in the treatment options of human Campylobacter infections of macrolide Erythromycin, ciprofloxacin, fluoroquinolone and tetracycline [34]. It also agrees with Moore et al., 2005) that people living in areas where there is high contact with farms animals and wild birds or chickens are the primary sources contributing to human Campylobacter infections [35].
And was also to agree with Rao et al., 2001, Who concluded that the age of Campylobacter infection in Sudan was in developing countries, that ranged in age of < 2-years- old children to 5 years, the diarrhea prevalence in children ≤ 5 years old, especially those living in rural areas [36]. The results were in disagreement with El-Sayed and Fahmy et al., 2005, in Egyptian Patients In Alexandria, the isolated C. jejuni from unselected children with diarrhea was apparent that the maximal incidence of Campylobacter infection occurs under the age of 3 years. And it was in contrast to the study by Lawson et al., 1999, study who found culture negative/and PCR-positive PCR-positive samples were found [37]. Our suggestion was Campylobacter growth in culture media were a failure or miss identified and possible of the PCR to detect and confirmed. The result were in disagrees with Murphy et al., 1994, in Thailand, the most isolates of C (Figures 4 & 5). jejuni were resistant to ciprofloxacin and nearly one third of isolates were resistant to Azithromycin, while those Khartoum State - Sudan isolates were sensitive to both antibiotics [38]. And were in disagreeing with Prats et al., 2000, in Barcelona, that isolates of Campylobacter jejuni were resistance to both ciprofloxacin and Nalidixic acid [39]. The finding were in disagreeing with Charvalos et al., 1995, on multidrug-resistant of Campylobacter jejuni to Erythromycin, Tetracycline, lactams and quinolones. But on the other hand, agree with the intermediate resistance to Gentamicin of Campylobacter jejuni [40]. And disagreement with previous findings in Australian Bacon et al., 1997, that tet (O) gene is the most common determinant conferring resistance to tetracycline, was analyzed its presence and localization in the tetracycline-resistant isolates [41]. In this study the finding was in agreed with Wagner et al., 2003, who detected Campylobacter jejuni from Germany; the isolates of Campylobacter jejuni were susceptible to Erythromycin, and Amoxicillin-clavulanic acid, but disagreed with resistant to gentamicin [42]. It was also in agreed with Moore et al., 2005, and Whyte et al., 2005, in that C. jejuni were sensitive to several antibiotics, including macrolides (especially Erythromycin), which have been used as the first-line therapy, and also quinolones such as ciprofloxacin [33,37].
Figure 4 Gel image of Campylobacter jejuni 161 bp size of PCR products from, Selected Hospitals in Khartoum state.
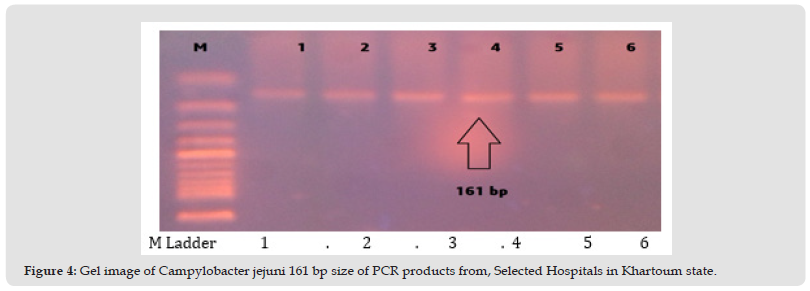
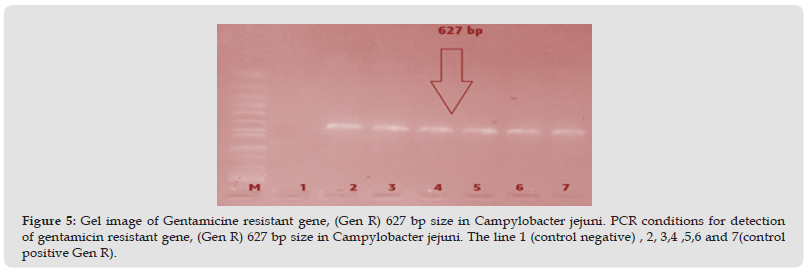
Figure 5 Gel image of Gentamicine resistant gene, (Gen R) 627 bp size in Campylobacter jejuni. PCR conditions for detection of gentamicin resistant gene, (Gen R) 627 bp size in Campylobacter jejuni. The line 1 (control negative) , 2, 3,4 ,5,6 and 7(control positive Gen R).
Campylobacter jejuni infection was the most common cause of campylo-bacterosis among younger children suffering from diarrheal disease seen in the selected hospitals in Khartoum state. In laboratory; Campylobacter jejuni was detected almost equal by conventional method and PCR techniques were consistent. The uncommon pathogen was Campylobacter coli which was not detected in selected hospitals in Khartoum state. The antibiotic treatment for Campylobacter jejuni showed that the isolates were susceptible to Erythromycin, Azithomycin, Tetracycline. Ciprofloxacin, and Amoxicillin /clavulanic acid. The resistance to Gentamicin by disc diffusion method was confirmed by PCR in gel electrophoresis at band size 627bp. In Gentamicin (Gen R), the Tetracycline and Ciprofloxacin resistance genes showed, were undetected. There were more Campylobacter jejuni seen in Khartoum North and Omdurman teaching hospitals than in other hospitals in the state. The area is limited to the banks of the river Blue Nile and White Nile, suggesting that there may be some sources of infection for children as well as contamination of the wider environment.
We would like to recommend the using a conventional method of isolation and identification of Campylobacter in routine microbiology laboratories. And application of molecular technique at central and regional reference laboratory in the surveillance of Campylobacter prevalence and antibiotic susceptibility pattern studies. The governmental state laboratories need to be supplied with human, lab equipment and materials in order to provide the service of isolation and sensitivity lab test for Campylobacter. We would like to recommend using a combination of multiplex molecular tests to reveal other pathogens causing diarrheal disease than Campylobacter. It also recommends encouraging microbiology labs to isolate, identify and do antimicrobial susceptibility testing to provide accurate data for antibiotic resistance surveillance. Its recommended to have a national Campylobacter reference lab capable of full facilities in identifying, characterization hand provides training of laboratory-based Campylobacter clinical specimen processing from human and animals and to assess the antibiotic susceptibility pattern of Campylobacter. Campylobacter control and prevention guidelines formulation from the ministry of health are on top demand to develop systematic therapeutic prescription of antibiotics for Campylobacter jejuni in children.
I would like to thank the staff of the Department of bacteriology in the National Health, for the exceptional the nice and pleasant place to work. Grateful to the molecular specialists for their Co-operation and making the place comfortable to for conducting this work.
All authors declared no conflict of interest.


