Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Md. Shakeel Ahmed1, Md. Zakir Hossain1, Md Rakibul Hassan1*, Zahirul Islam1, Md. Fazle Rabby2, Mahabubul Mowla2, Kishor Kumar Barua1, Kuldeep Sharma1, Md. Ziaur Rahman1, Md. Meherab Hossain1 and Hasan Rabbi1
Received: November 23, 2022; Published: December 06, 2022
*Corresponding author: Md Rakibul Hassan, Bangladesh Institute of Tropical and Infectious Diseases Fouzderhat, Chittagong, Bangladesh
DOI: 10.26717/BJSTR.2022.47.007506
Background: Rapid and reliable detection of SARS-CoV-2 by reverse transcriptase quantitative real-time polymerase chain reaction (RT-qPCR) plays a pivotal role in the timely identification of cases and targeted measures to prevent transmission of COVID-19. Large-scale screening of incoming travelers and patients with symptoms or those with a history of exposure is critical to managing the COVID-19 pandemic for any country. However, the extra resources, manpower, and infrastructure required for such large-scale testing are limited in many low-income countries. Specimen pooling for COVID-19 testing in low-prevalence settings is now recognized as an effective strategy to save scarce resources. The study aimed to evaluate the pool testing of clinical samples through RT-PCR compared with the individual RT-PCR test.
Method: We assessed a pooled specimen testing approach on directly lysed nasopharyngeal specimens collected from suspected COVID-19 patients in Bangladesh using a commercial SARS-CoV-2 detection kit.
Results: A total of 3985 samples were tested individually or in the pooled testing, where each pool was comprised of five specimens. An individual RT-PCR test result was taken as a gold standard to compare the result and used to calculate the diagnostic proficiency of pool testing. Out of 797 pools, 714 were made up of 3570 samples, were negative; and 79 pools, made up of 395 samples, were positive. The sensitivity and specificity of pooled specimen testing were 96.9 % (95% CI, 95.1 to 98.1) and 100%, 100%, and 99.9 % (95% CI, 99.9 to 99.9), respectively, with reference to individual test results. The overall agreement was 99.9 % (95% CI, 99.9 to 99.9: kappa = 0.993). With only 10% of pools being positive for SARS-CoV-2 in our setting, individual re-testing would have been required for only 395 samples, saving a total of 2777 (77.8%) PCR tests.
Conclusion: Our results suggest that applying directly lysed specimens instead of conventional RNA extraction kits and pooling up to 5 samples for COVID-19 testing individual laboratories can significantly reduce tests cost, save limited resources, and expand test capacity without compromising the quality of the test.
Coronavirus disease - 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) first emerged in December 2019 in Wuhan, China [1]. The novel virus, SARS-CoV-2, is a member of the human coronavirus (HCoV) family that cause respiratory tract infections [2]. It has challenged every aspect of human lives and overwhelmed the national healthcare systems of the affected countries worldwide. Although several vaccines have now been approved, and a massive vaccination campaign is ongoing in many countries, it will still take a long time to vaccinate a significant proportion of the world’s population. Furthermore, there is a considerable shortage in the supply of vaccines in low-income countries. Therefore, testing, tracing, and isolation remains the primary strategy to prevent widespread virus transmission in many countries. However, testing thousands of people in a shorter period is a daunting task that demands extensive resources and expertise, which may be limiting in many countries of the world [2,3]. To maximize testing capacity in the face of the shortage of laboratory equipment, reagents, and resources, alternative strategies such as the pooling of specimens are needed to reduce the cost and labor associated with the test and increase the throughput of the test. [4-6]. Dorfman [7] introduced a simplified pooling strategy in which all the pools tested contained the same number of specimens.
If a specific pool tests positive, all of the specimens belonging to the pool are re-tested to identify the positive samples within the pool. Reverse transcriptase quantitative real-time polymerase chain reaction (RT-qPCR) is a highly sensitive tool. It has previously been used efficiently in combination with sample pooling strategies for the surveillance of HIV and hepatitis B and C viruses when the prevalence is under 30% and when it has been proven cost-effective [8,9]. Pool testing has also been evaluated for COVID-19 in several recent studies [8,9]. Because in a low prevalence setting, only a few of the tested pools would turn positive and need to be re-tested, the sample pooling strategy is expected to save cost, reagents, labor, and instrument time of tests and improve the turnaround time of negative test results. For the same reason, pool testing has also been recommended by the Centers for Disease Control and Prevention (CDC) and the Federal Drug Agency (FDA) to screen asymptomatic individuals. However, most studies on specimen pooling have been conducted in developed countries, and the performance characteristics of pool testing using a direct sample lysis protocol have not been evaluated. In this study, we have applied Novel Coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR-Fluorescence Probing) (Sansure Biotech Inc.) that uses an RNA extraction free, direct RT-PCR approach on lysed cells harvested from nasopharyngeal swab (NPS) specimens. In response to the emergency arising from the COVID-19 pandemic, and the shortage of the SARS-CoV-2 testing kits in different testing facilities around the country [10,11], we evaluated the performance of sample pooling approach concerning the results of individual testing for the detection of SARS-CoV-2 by using the Sansure Biotech SARS-CoV-2 RT-PCR Kit.
Sample Collection from Suspected Covid-19 Patients
Clinical samples (Nasopharyngeal swabs) were collected from the Bangladesh Institute of Tropical and Infectious Disease (BITID), Fouzderhat, Chittagong. Nasopharyngeal (NPS) was collected in a 2mL sample storage buffer. Collected specimens to be tested were immediately processed or stored at 40 C for testing within 24 hours of sample collection. Specimens that could not be tested within 24 hours were stored at -200C for ten days. This study followed the principles of the Declaration of Helsinki of the World Medical Association. Ethical permission was obtained from the ethical committee of the Bangladesh Institute of Tropical and Infectious Diseases (BITID/EP-39/2021).
RNA Extraction Through Direct Lysis
RNA was extracted using the one-stop method of sample lysis using lysis buffer from Sansure Biotech, China. Briefly, 10 μL of sample in sample storage buffer was taken into a 1.5 ml micro-centrifuge tube followed by 10 μL addition of sample lysis buffer (Sansure Biotech, China). Incubate the mixture at room temperature for 10 minutes. Keep the tubes containing cell lysate at -20 ˚C until qRTPCR is performed.
Sample Preparation and RT-qPCR
10 μL of NPS specimen was transferred to a 1.5 ml micro-centrifuge tube and added 10 μL sample lysis buffer (Sansure Biotech, China). The mixture was incubated at room temperature for 10 minutes and centrifuged at 8000 rpm for 3 minutes. Tubes containing cell lysate were stored at 20 ˚C until RT-qPCR was performed. A novel Coronavirus (2019-nCoV) nucleic acid diagnostic kit (PCR-Fluorescence Probing from Sansure Biotech, China) was used to detect the ORF1ab and N genes of SARS-CoV-2 RNA qualitatively. 20 μl of cell lysate was added to 30 μl of 2019-nCoV-PCR master mix (2019-nCoV-PCR Mix + 2019-nCoV-PCR-Enzyme Mix) to amplify on Bio-rad CFX96 touch real-time RT-PCR detection system (Bio-Rad Laboratories).
Initial Optimization of Pool Testing Approach
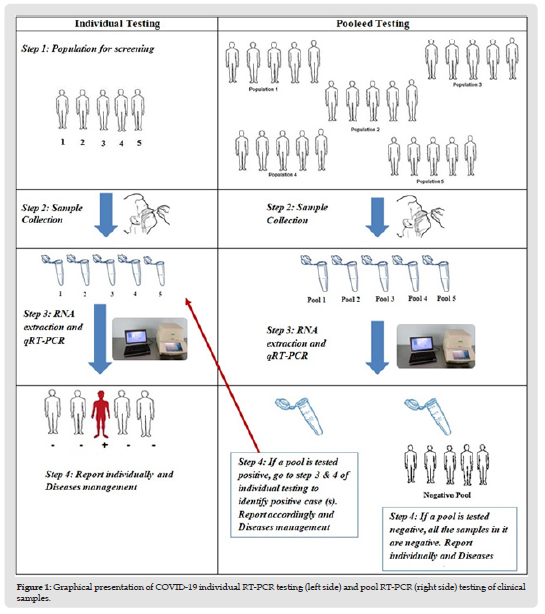
A total of 20 pools were established using 100 NPS specimens (5 specimens/pool) that were previously tested positive or negative for COVID-19 (87 negative and 13 positive clinical specimens). An aliquot (10 μL) of the specimen was transferred from 5 specimens into a 1.5 ml micro-centrifuge tube to prepare a pool with a final volume of 50 μL. Each pool was then subjected to centrifugation at 8000 rpm for 5 minutes. 40 μL supernatant was discarded from each pool, and the remaining supernatant, including the pellet, was thoroughly mixed with 10 μL of the sample lysis buffer (Sansure Biotech, China). The mixture was incubated at room temperature for 10 minutes and subjected to RT-qPCR analysis for SARS-CoV-2 according to the manufacturer’s instructions (Sansure Biotech, China). A graphical exhibition of how we executed the pool testing strategy is described in (Figure 1).
Figure 1 Graphical presentation of COVID-19 individual RT-PCR testing (left side) and pool RT-PCR (right side) testing of clinical samples.
Evaluation of the Pool-Testing Approach
A total of 3985 were used in 797 pools (each pool with five specimens) in parallel with their individual assessment. Samples for pooling were selected randomly. RT-PCR cycle threshold (Ct) values of 38 or higher were considered negative results and applied to individual and pool testing. Diagnostic performances like sensitivity, specificity, positive predictive value, negative predictive value, and relative efficiency were calculated by comparing pooledtest data with the individual test results.
Data Analysis Technique
SPSS (Statistical Package for Social Science) for Windows version 23 software was used for the analyses. Continuous variables were reported as the means ± SD, and categorical variables were notified as percentages. Student’s t-test was performed to compare continuous variables. For each test, sensitivity, specificity, positive predictive value, negative predictive value, and accuracy were calculated, and 95% confidence intervals were estimated. The individual RT-PCR test result was considered the gold standard for analytical comparison. Statistical significance was defined as p < 0.05, and the confidence interval was set at 95%.
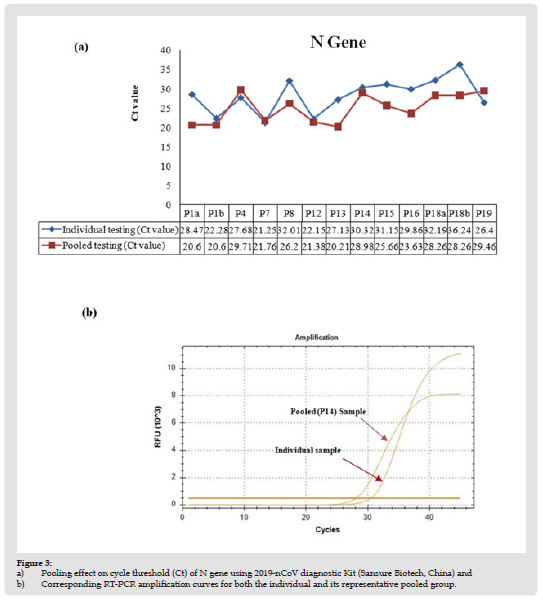
For initial testing, a total of 20 pools (5 specimens/pool) were established from 100 residual NPS specimens after Sansure SARS-CoV-2 RT-qPCR individually tested them. Among the pools, nine contained all negative samples, two contained 2 SARS-CoV-2 positive specimens each, and nine pools contained 1 SARS-CoV-2 positive sample each (Table 1). Positive specimens with a different range of Ct values were chosen. Ct values for SARS-CoV-2 ORF-1AB and N gene obtained by pool testing and individual testing were compared (Figures 2 & 3). Qualitative test results were 100% concordant. All negative pools tested negative, and all positive pools tested positive. Surprisingly, for pools containing only one positive sample, the average CT value for ORF-1AB is higher (ΔCt = 4.5± 3.4; p=0.0042 by paired t-test) in individual tests than in the pooled tests. However, the CT value of the N gene was not significantly different (ΔCt = 2.3 ± 3.8; p = 0.106 by paired t-test) between the two. These results suggest no loss of analytical sensitivity because of sample pooling.
Note: (P1 ...…. P20) = (Pool 1...Pool 20)
Figure 2 a) Pooling effect on cycle threshold (Ct) of ORF-1AB gene using 2019-nCoV diagnostic Kit (Sansure Biotech, China) and b) Corresponding RT-PCR amplification curves for both the individual and its representative pooled group.
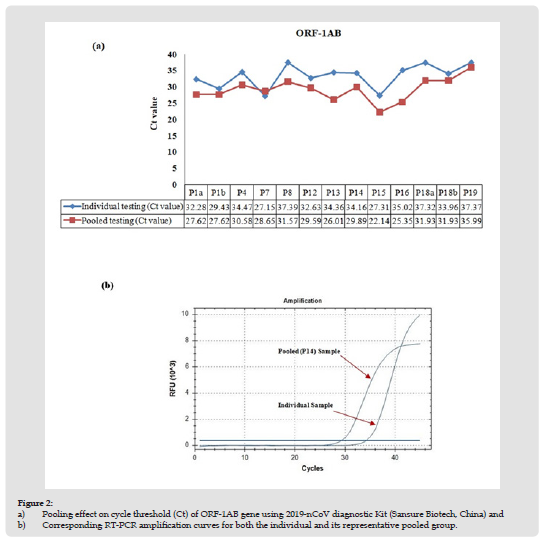
Figure 3 a) Pooling effect on cycle threshold (Ct) of N gene using 2019-nCoV diagnostic Kit (Sansure Biotech, China) and b) Corresponding RT-PCR amplification curves for both the individual and its representative pooled group.
For prospective evaluation, 3985 samples were analyzed from two different sites in Bangladesh. All the samples were analyzed individually and also, in parallel, pooled into 797 groups of samples (each pool contained five specimens). Ninety-five samples (2.38%) were found positive. We found that 714 pools, made up of 3570 samples, were negative, and nine pools, made up of 395 samples, were positive. Four pools comprising 20 samples were invalid. Therefore, our pooling strategy would have saved 2777 (77.8%) PCR tests. According to the Dorfman pooling method, for any given assignment of p (prevalence rate) and n (pool size), the expected Dorfman optimal efficiency is calculated as
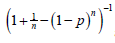
Based on our study and the prevalence of SARS-CoV-2 in our patient population, implementing a pooled sample testing approach to detect SARS-CoV-2 by RT-PCR would bring significant cost-savings and reduction of workload because of the much lower number of PCR tests necessary with the specimen pooling approach. Furthermore, the pool testing approach can significantly improve the turnaround time for reporting negative results. Such a rapid diagnosis strategy helps to track down the early community transmission of SARS-CoV-2 while appropriate and adequate infection control measurements can be implemented to reduce transmission of the disease [12,13]. Sample pool testing has been practiced previously for HIV and Influenza virus surveillance, and recently many reports have justified the pool testing for COVID-19 screening [14- 16,4-6]. In this study, we provide additional data on sample pooling for COVID-19 testing generated in the context of a resource-limited country. Furthermore, we have provided data on an extraction-free protocol that provides additional cost savings and reduces the analytical time of the test.
Approximately 700 COVID-19 tests are performed in our laboratory daily to screen flight passengers. Tests are typically requested to meet pre-travel requirements; therefore, many samples are expected to be negative. In this study, with a positivity rate of only 2.38%, only a tiny fraction of sample pools required re-testing. Furthermore, our results show > 95% sensitivity and specificity against individual test results. Post-implementation, pool testing has significantly reduced the backlog of pending COVID-19 tests in our laboratory and improved the turn-around reporting time (data not shown). [17] However, it should be noted that sample pooling may not be cost-effective or convenient for COVID-19 testing in populations with higher positivity rates. Another limitation of our study is that test results were generated in a single center. Additional data from other test centers in Bangladesh or other countries using the same RT-qPCR test kit may further confirm the validity of pool-test results obtained with the Sensure RT-qPCR kit. In conclusion, we present here a simple, rapid, and inexpensive pool-testing approach for SARS-CoV-2 detection that could provide significant benefit over the standard approach, in a low prevalence setting, in terms of cost, labor, and test capacity. Our approach is beneficial for countries like Bangladesh, where the rate of COVID-19 PCR testing is far fewer than the developed countries because of the limitations of technology, expertise, and resources. In the wake of the COVID-19 pandemic, Bangladesh has significantly expanded its RT-qPCR facilities [18]. However, test capacity per laboratory is still limited because of a shortage of test reagents and workforce. Implementation of a pool testing approach in these laboratories, depending on their rate of SARS-CoV-2 PCR positivity, may assist the country in managing the COVID-19 pandemic more efficiently.
Md. Shakeel Ahmed was the principle investigator responsible for the project administration and supervision of the study. Md. Zakir Hossain was responsible for the laboratory resources and supervision of the laboratory staff. Zahirul Islam’s role was laboratory investigation, methodology, and validation. Md. Fazlee Rabby, Mahabub Mowla, Kishor Barua, and Kuldeep Sharma assisted in patient specimen collections and laboratory methodology. Rakibul Hassan’s role was an investigation, validation, and formal data analysis, responsible for the original draft writing and editing of the manuscript. All authors approved the final version of the manuscript.
The study received no external or internal funding.
The authors do not have a commercial or other association that might pose a conflict of interest.
The authors thank Rodolphe Merièux Laboratory, Fouzderhat, Chittagong, for providing laboratory facilities and technical support throughout the study.


