Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Sotiris Raptis1, Christos Ilioudis2, Vasiliki Softa1 and Kiki Theodorou1*
Received: November 15, 2022; Published: December 02, 2022
*Corresponding author: Kiki Theodorou, Medical Physics Department, Medical School, University of Thessaly, Greece
DOI: 10.26717/BJSTR.2022.47.007497
Radiomics is a new term in radiology that refers to the extraction of a large number of quantitative information from medical pictures. Artificial intelligence (AI) is a wide term for a collection of advanced computing algorithms that, in essence, analyze patterns in data to make predictions on previously unknown data sets. Because of its superior ability to handle large amounts of data as compared to traditional statistical methods, it can be combined with AI. We performed a comprehensive review of the available literature on studies investigating the role of radiomics and radiogenomics models in order to improve the prediction treatment response in non-small-cell lung cancer (NSCLC). The basic goal of these fields, taken together, is to extract and analyze as much useful hidden quantitative data as possible for decision support. Radiomics and Artificial Intelligence have been used extensively to characterize lung malignancies. Furthermore, it has been successfully used to forecast side effects such as radiation- and immunotherapy-induced pneumonitis, as well as to distinguish lung injury from recurrence. Our research interest in this paper is to provide an update on the current status of the use of radiomics and artificial intelligence in lung cancer, detecting the existing gap in order to develop diagnostic, predictive, or prognostic models for outcomes of interest.
Keywords: Artificial Intelligence; Radiomics; Non-Small-Cell Lung; Precision Medicine
Abbreviations: AI: Artificial Intelligence; WHO: Word Health Organization; CT: Computed Tomography; MRI: Magnetic Resonance Imaging; PET: Positron Emission Tomography; ML: Machine Learning; DL: Deep Learning; NSCLC: Non-Small-Cell Lung Carcinoma; ROI: Region of Interest; IBSI: Image Biomarker Standardisation Initiative; DICOM: Digital Imaging and Communication in Medicine; MSE: Mean Squared Error; WEKA: Waikato Environment for Knowledge Analysis; RP: Radiation Pneumonitis
Radiomics is a new term for the area of radiology, derived from a combination of “radio,” which refers to medical imaging, and “omics,” which refers to the numerous fields such as genomics and proteomics that help us comprehend diverse medical diseases. Given the massive efforts to meet the unmet clinical need that still exists, it is predicted to have a significant impact on clinical practice in the near future in the domain of lung cancer. According to Word Health Organization (WHO), cancer is a leading cause of death worldwide, accounting for nearly 10 million deaths in 2020 [1]. Lung cancer is scientifically proven to be the most common cause of cancer-related death worldwide; advances in early, potentially treatable diagnosis would have a significant impact on human health. As a result, lung cancer became the first malignancy for which radiomics have been used in clinical trials and so we can expect that Artificial Intelligence (AI) will speed the clinical translation of lung cancer radiomics by attaining fully automated patient prediction. The goal is to extract quantitative and actionable information from images such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) that are not easily visible or quantifiable with the specialist-radiologist eye, in order to build a model assessing clinical outcomes, including diagnostic, prognostic, and predictive perspectives, to predict outcomes.
Today, Artificial Intelligence provides a plethora of essential tools for intelligent data analysis, which may be used to solve a variety of medical difficulties, including diagnostic ones. First of all, we have to explain the difference between the main terms of Artificial Intelligence (AI), Machine Learning (ML) and Deep Learning (DL). Artificial intelligence is the process of incorporating human intellect into machines via a set of rules such as algorithms. Machine Learning is the process that allows a machine to learn on its own and develop as a result of its experiences without having to be explicitly programmed, so ML is a subset or application of AI. Deep Learning is a subset of Machine Learning that uses Neural Networks (similar to the neurons in our brain) to replicate human brain-like behavior. DL algorithms concentrate on information processing patterns mechanisms in order to find patterns in the same way that our human brain does and classify data accordingly.
When compared to ML, DL works with bigger volumes of data and the prediction method is self-administered by computers. The most common kind of lung cancer is non-small-cell lung carcinoma (NSCLC), which accounts for more than 80 percent of all lung can cer diagnoses. In this paper, we summarize the present state and assess the scientific and reporting quality of radiomics research in the prediction of treatment response in non-small-cell lung cancer. There are numerous potential radiomic indicators of therapy response in lung cancer described in the included reports; however there is a gap of clinical evaluation between the current statuses of the relevant studies. Our study focuses on the deployment of standardized features and software in a prospective scenario, as well as external validation. Radiomics, being a new and evolving medical instrument, continues to face problems that prevent its widespread usage in clinical practice. Despite these problems, there is a lot of research going on in the sector, which might lead to this new technological approach becoming an important therapeutic tool in the future.
Radiomics
Radiomics is the extraction of usable data from medical imaging that has been used in oncology to enhance diagnosis, prognosis, and clinical decision support with the objective of providing precision medicine [2]. This analysis might be viewed as a virtual biopsy tool, with the ability to detect and determine tumor phenotypes, thanks to technical advancements in AI, using various imaging modalities, such as CT, PET CT, and MRI. Its features derived from MRI can be an important tool even in limited patient population [3]. The workflow is interdisciplinary, comprising radiologists, data scientists, and imaging scientists, and it involves tumor segmentation, image preprocessing, feature extraction, model construction, and validation in a sequential procedure. The distribution of signal intensities and spatial connection of a three- dimensional grid (voxel) within a region of interest (ROI) are often described by extracted features. The idea is that imaging data can provide information about tumor biology, behavior, and pathogenesis that isn’t visible to existing radiologic and clinical interpretation [4]. Despite the fact that many of the principles of image feature extraction have been around for decades, the field’s research output has exploded, according to PubMed (www.pubmed.gov), with over 1500 articles using the phrase radiomics in 2020. It can be used with computed tomography, magnetic resonance imaging (MRI), X-ray, positron- emission tomography, and ultrasound, among other imaging modalities. This procedure begins with picture capture, the parameters and protocol must be carefully considered. A consistent set of picture acquisition procedures is desirable to extract features in a stable and repeatable way (Figure 1).
After the data has been collected and arranged, the regions of interest on the pictures to be studied are segmented. The ROI designates the area from which radiomics characteristics will be retrieved, and manual or semiautomatic segmentation by a competent expert (e.g., a doctor) is presently the radiomics analysis’ rate-limiting phase. The process of calculating features is lengthy and complicated, resulting in inaccurate reporting of methodological data (e.g., texture matrix design choices and gray-level discretization approaches). A previous study found that higher order radiomics features were related with substantial changes in features retrieved using different toolboxes, whereas histogram-based features were the most repeatable [5]. The Image Biomarker Standardisation Initiative (IBSI) has created a standardized radiomics procedure, which is shown in (Figure 2). The IBSI intends to standardize both feature calculation and the image processing tasks that must be completed before feature extraction. A basic digital phantom was created for this purpose and utilized in Phase 1 of the IBSI to standardize the computation of several variables from many categories, including morphologic, local intensity, and statistical. A collection of CT scans from a lung cancer patient was employed in Phase 2 of the IBSI to standardize image processing stages utilizing five distinct parameter combinations, including volumetric techniques (2D versus 3D), picture interpolation, resegmentation, and discretization methods [6]. Imaging data that is suitable for the research and meets the inclusion and exclusion criteria should be explicitly stated. To decrease unjustified confounders and noise, standard imaging techniques (i.e., those that employ the same vendor or scanner settings for all samples) might be used. Images should be anonymised to eliminate patient sensitive metadata after a cohort has been established. To prevent losing potentially valuable picture elements, images should be exported as Digital Imaging and Communication in Medicine (DICOM) files using a lossless compressed format.
Artificial Intelligence
Why AI in imaging? First of all, AI is non-invasive and easy to repeat and this is important as tumors are spatially and temporally heterogeneous, in addition there is the need for innovation. The fundamental rationale for AI’s use in radiomics is that it is better at handling large amounts of data than traditional statistical methods. The majority of AI algorithms are employed to solve classification problems. These algorithms basically analyze patterns in the data presented and then make predictions on unknown data sets to see if the patterns are right. AI algorithms are capable of analyzing not just the quantitative data provided by predefined or hand-crafted radiomic features, but also of analyzing images directly in order to generate its own characteristics. To improve generalizability, these methods can be paired with meta-classifiers or ensemble approaches like adaptive boosting and bootstrap aggregation. There are also additional ensemble learning approaches that combine many algorithms, such as k-nearest neighbors, naive Bayes, and tree algorithms, which are poor classifiers [7]. This advanced subset of AI is called deep learning. According to Giovanni L. F. da Silva et al [4], deep learning methods were useful in recognition of small lung nodules on CT, with an accuracy of 97,6%. Deep learning is a very prominent and advanced subset of AI [8]. These systems can also do segmentation tasks on their own, requiring no human intervention.
As we mentioned it in the introductory of this paper - artificial intelligence can help in many ways in the healthcare sector - let it be diagnostic, therapy, drug development, patient workflow management or remote diagnosis. One of the most popular machine learning approach to tackle these problems is deep learning. The application of deep learning techniques and especially Neural networks for medical image segmentation received a great interest due to their ability to learn and process large amounts of data in a fast and accurate manners. What are Neural Networks? At a first sight we can see neural networks as black boxes where we are putting in some data to the input and on the other end, we will get an analysis coming of the box - let it be segmentation, or a classification for example. If we open this box we will see (in case of a fully connected network) plenty of neurons and connections between them. Layers of neurons stacked on each other (Figure 3). Origins of neural networks were inspired by the human brain itself (however the network design used now is very loosely connected to it). These networks consist of neurons where the connection between them is modeled with weights. As we are moving forward in the network each output of a neuron calculated as the following:
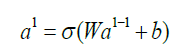
Where al is the output of the neuron at the “l”th layer, al−1 is the output of the previous layer called activations, W are the weights and b is called bias. Step by step as we are moving forward we will reach the end of the network, the output layer - the output of this layer will be the result provided by the network. At this point the network may have some error - the output is different from what is expected. To measure the difference we are using a mathematical function which is called loss function. Depending on the task we are trying to solve we may use different loss functions, and we may modify this loss function during the training. One good example for a loss function is a simple mean squared error (called MSE):
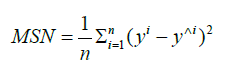
Where n is the number of examples seen by the network, y is the expected output and y^ is the actual output. Now that we have numerical information about the error of the network we can try to mitigate it and optimize the network to for example in case image classification - minimize it. The optimization method used in a neural network in deep learning is called backpropagation. We are propagating the error back from the output to the input - meanwhile updating the weights and biases in each neuron. If you are not into math - all you need to understand is that we are calculating how much a neuron is contributing to the final error and based on this number we are modifying its weight so the loss function will be optimized.
Tracing the Gap
Because of variances in patient demographic, cancer stage, treatment mode, and radiomics workflow technique, there is a lot of variability between current studies. The management of the patients would be different if the prognosis of the patients could be anticipated before any intervention or treatment. Precision medicine is the term for this. Except for a few parameters like size and volume, imaging data sets are mainly analyzed visually or qualitatively in traditional radiology practice. This method not only introduces intra- and interobserver variability, but it also ignores a significant amount of concealed information in medical imaging. The lesion is often manually demarcated by an expert radiologist or radiation oncologist in radiomics studies of lung cancer. Automatic or semi-automatic segmentation, on the other hand, improves repeatability and yields more stable characteristics than manual segmentation. Because lung nodules might be small and appear similar to other structures in the lung, such as blood vessels or benign processes like localized organizing pneumonia, pulmonary imaging provides distinct problems to both radiologists and radiomics systems. As a result, CT-based lung cancer screening has a significant false-positive rate [9].
In positron-emission tomography (PET), when picture noise affects segmentation, a relative threshold method or variational approaches can be used to semi-automatically define lung tumors by attempting to utilize gradient differences between the foreground lesion and the background [10]. The basic goal of radiomics is to extract as much and meaningful hidden objective data as feasible for use in decision support using either standard or sophisticated imaging techniques [11]. However, the limited replication potential of most current research can be partly blamed for the fact that AIbased techniques have yet to be incorporated into normal practice. For medical imaging segmentation, there are a number of commercially accessible software solutions. As a result, discrepancies in observers and software might lead to considerable variances in feature values. In radiomics analysis, interobserver variability has been somewhat addressed by employing characteristics that do not differ significantly among observers. This method, however, may result in the omission of characteristics that may help the predictive/ prognostic/diagnostic model.
Deep learning approaches have recently been used to construct auto-segmentation systems, and the results have been encouraging [12]. When it comes to software, the first decision to make is whether to utilize commercial or nonprofit software. Noncommercial apps are usually free, constantly changing, and reflect current research trends. Commercial programs aren’t free, but they’re more likely to be stable, come with technical assistance, and be a “black box”. Using free software applications that allow the export of radioactive properties with a graphical user interface is a slightly more complex method. IBEX [13], LIFEx [14], and PyRadiomics [15] are some of the most widely used applications for exporting handcrafted features. However, radiologists should exercise caution when using these software programs because the pipeline is not well established in such programs, and there are numerous parameters to be dealt with, such as establishing discretization levels, normalization approach, re-sampling, and clearing non-radiomic data from the final feature table. The aforementioned gap comes to fill our familiarity with coding skills. Following feature selection, interesting models may be created using a variety of machine learning methods. Various modeling methods, ranging from basic decision trees or logistic regressions to more complicated random forests or Bayesian networks, are implemented in software such as R https:// www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical- computing), MATLAB ttps://www.mathworks.com/), and scikit-learn (https://scikit-learn.org/stable/).
The most popular platforms for this purpose are the various Python platforms, which have huge libraries for both handmade and deep feature extraction. Additionally, the Waikato environment for knowledge analysis (WEKA) [16], and Deep Learning Studio (https://deepcognition.ai/) are some software applications that can be used for this purpose, WEKA is capable of performing various ML tasks, however, its visual capabilities are restricted and poor until it is integrated with other settings. Currently, a wide range of acquisition methods are in use. Furthermore, numerous vendors offer a variety of image reconstruction methods, which are tailored to the needs of each institution. This is a problem not only on a multi- institutional scale, but also within a single institution. Although it is often overlooked or ignored in visual analysis, the use of various acquisition and image processing techniques may have a significant impact in deep learning because it is a pixel or voxel- level process that can affect image noise and texture, potentially reflecting a different underlying pathology [17].
Several research organizations have looked into using AI to predict treatment response, but it has yet to be put into clinical practice. Choosing the best method for model creation is a hot topic in academia right now. Recent works have been reported on automating the process of picking the optimal algorithm for a particular dataset [18]. Although algorithm selection appears to be random in the literature, the optimum approach would be algorithm selection based on several experiments. From a practical standpoint, we must realize that it is impossible to standardize all image capture procedures. Our major objective, on the other hand, should be to develop the optimal technological pipeline for creating the most stable and accurate AI models that can be applied to pictures collected via various protocols.
Radiomics is now regarded as a purely academic field. The aim of this narrative review is to report the recent literature, and to provide an update on the current status of the use of radiomics and artificial intelligence in Lung Cancer in order to develop diagnostic, predictive, or prognostic models for outcomes of interest. Before being used as a therapeutic decision-making tool to support individualised therapy for patients with lung cancer, promising AI models must be externally verified and their effect studied within the clinical pathway. The results must be confirmed using separate data sets in order to be accepted in the clinical arena. Techniques for internal validation can be applied. K-fold, leave-one-out cross-validation, and hold-out are the most frequent internal validation procedures found in the literature. The most essential issue to address in internal validation is the possibility of feature selection algorithm leaking across the data, which might lead to unduly optimistic conclusions. Conventional statistical approaches may be used to compare the validation performance of our AI systems. Despite the fact that the number of relative papers has increased exponentially, normal clinical adoption has yet to emerge [19]. Noncompliance with machine learning best practices, standardization of its process, and unambiguous reporting of research methodology are all major roadblocks. Only then may models be evaluated using external real-world data, such as multivendor pictures and a range of acquisition procedures, ideally prospectively.
In order to overcome these obstacles, data curation and quality, as well as suitable sample numbers, are essential. As researchers have shown, these systems can outclass humans while performing medical image analysis, they have been created to assist in improving predictive analytics and diagnostic performance, specifically to improve their accuracy and capacity to enable personalized decision-making [20]. Curation of enormous datasets, on the other hand, requires a lot of time and effort, and gathering enough data from many institutions might be difficult. These issues can be addressed with the use of data exchange. Finally, while artificial intelligence is essentially a data-driven endeavor, a better understanding of the physiological significance of any generated radiomic signatures is necessary before the findings are widely accepted.
As a step further in our research, the treatment of lung cancer varies depending on the histologic type, cancer stage and the personal status; it includes surgery, chemotherapy and radiotherapy. The most common side effect of radiotherapy lung cancer is Radiation Pneumonitis (RP) as a symptomatic toxicity caused by an inflammatory response to radiation. The likelihood of symptomatic RP could be reduced if it could be predicted early in the radiation therapy regimen. Radiomics can provide quantitative features from medical imaging [21]. In relation to the above, we develop a model, using Artificial Intelligence and emerging technology which allows us to have access to far more data than was ever previously available to it. Moreover, our laboratory will analyze the texture characteristics of computed tomography as possible prognostic factors of RP. The high dimensionality of radiomic datasets is a major issue. Our proposed method in data problems is the synthetic minority oversampling technique in order to create a balanced dataset. We leverage suitable hardware and open-source software framework by Pytorch and other software solutions like DNN library.
There are no conflicts of interest to declare.


