Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Raviswamy GH Math1, Harish Holla2, Doddabasappa Talimarada2, Vikas Kumar3, Yoshita S4, Veena Prasad4, Suwanand Deshmukh1, Savithri Bhat4 and Dhananjay D Shinde*5
Received: October 08, 2022; Published: October 21, 2022
*Corresponding author: Dhananjay D Shinde, Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, NE, USA
DOI: 10.26717/BJSTR.2022.46.007399
Metabolomics and lipidomics analysis target a huge number of intermediate and end products of biological fluids, tissues, and cells. Bacopa monnieri, commonly known as Brahmi is a nootropic plant that widely used in Ayurveda treatment for improving memory and reducing anxiety. Some research suggests that it might also protect brain cells from chemicals involved in alleviating symptoms of neurological disorder such as Alzheimer’s disease. To evaluate the chemical composition of the bioactive metabolites and lipids, ultra-high-performance liquid chromatography coupled with electrospray ionization hybrid linear trap quadrupole-orbitrap mass spectrometry (UHPLC-ESI/LTQ Orbitrap-MS) was applied. Along with UHPLC-HRMS, bioinformatics tools such as Compound discoverer and Lipid search software packages were used for the identification and quantification of metabolites and lipids in roots, stem, and leaves of Bacopa plant. Metabolites and lipids were characterized based on highly resolved precursor ions and product ions. A pool of labeled reference standards for lipids was used to identify the precursor ions and fragmentation pattern. The classes of identified compounds included glycerophospholipids, glycerolipids, sphingolipids, mono-, di-, and triglycerides, saponins, bacosides etc. Developed methods were applied successfully for the relative quantitation and differential expression of these biomolecules in roots, stem, and leaves of Bacopa plant.
Keywords: Bacopa Monnieri; LC-HRMS; Lipidomics; Metabolomics
Abbreviations: BM: Bacopa Monnieri; BME: BM Extract; PD: Parkinson’s Disease; ROS: Reactive Oxygen Species; BBB: Blood-Brain Barrier; GKVK: Gandhi Krishi Vigyana Kendra; BHT: Butylated Hydroxytoluene; UHPLC: Ultra High-Performance Liquid Chromatography; LCHRMS: Liquid Chromatography-High Resolution Mass Spectrometry
Bacopa monnieri (BM) also known by the common names waterhyssop (Roodenrys, et al. [1,2]) brahmi (Sivaramakrishna, et al. [2]), thyme-leafed gratiola, herb of grace (Sivaramakrishna, et al. [2]), and Indian pennywort (Roodenrys, et al. [1]) is a perennial, creeping herb native to the wetlands of southern and Eastern India, Australia, Europe, Africa, Asia, and North and South America. Both traditional Ayurveda medicines and food supplements have been using BM since ancient times for improving memory and treatment of neurological disorders. Several medicinal applications of BM and its extracts have been reported as the significant anti-depressant, anti-anxiety, anti-convulsant, anti-cancer, anti-inflammatory, antioxidant, anti-bacterial, anti-diarrheal, anti-hypertensive, analgesic and anti-toxicity activity [3-5]. Salvia lavandulaefolia (Spanish sage) and Salvia officinalis (common sage) are being used for improving memory in Europe since the 16th century (Akhondzadeh, et al. [6,7]) whereas another traditional Ayurvedic remedy; Centella asiatica (Asiatic pennywort), is commonly used in combination with milk to improve memory. Parts of other herbs such as roots of Withania somnifera, are also used in Ayurveda as a rejuvenative tonic for enhancing memory. In preliminary clinical research, it is observed that BM may improve cognition (Kongkeaw, et al. [8,3,9]).
Formation of transporter of thyroxine and vitamin A such as homotetrameric plasma protein transthyretin (TTR) fibrils are restricted due to the use of BM extract (BME) in human trials, highlighting the neuroprotective role of TTR in Alzheimer’s disease. After BME treatment, TTR amyloidogenesis is inhibited by attenuating the disassembly of tetramers into monomers [10]. In animal models exposed to BM, the neuro-protective effects were observed via regulating neuroinflammation against Parkinson’s disease (PD) due to the reduced levels of pro-inflammatory cytokines together with decreased levels of α-synuclein and reactive oxygen species (ROS) generation. These results suggest that BM can limit inflammation in the different areas of brain, thus, offers a promising source of novel therapeutics for the treatment of many CNS disorders [11]. The synergic effect of dietary polyphenolic compounds on neurocognitive function and their use in nutritional intervention studies of the brain are well established [12]. Ayurveda practitioners routinel use BM to treat the various ailments such as memory loss, inflammation, epilepsy, fever, and asthma. Numerous studies suggested that bacosides; prominent bioactive components of BM protect the brain against oxidative damage and age-related cognitive deterioration. Treatment with bacosides prevents Aß aggregation and formation of fibrils (Holcomb, et al. [13]) as well as it helps to protect neurons against Aβ-induced toxicity [14].
The best characterized phytochemicals in BM are dammaranetype triterpenoid saponins known as bacosides, with jujubogenin or pseudo-jujubogenin moieties as aglycone units [10]. Bacosides (Bac), the key ingredients present in BM and BME are nonpolar glycosides (Chakravarty, et al. [15]). These bacosides though a simple lipid-mediated passive diffusion [16] have ability to cross the blood-brain barrier (BBB). The radiopharmaceuticals biodistribution studies strongly affirm the bioavailability of bacosides in the brain. It is well established in previous studies that the combined doses of bac I and bac II were synergistic and reduced the viability and proliferation of the four breast cancer cell lines when administered each below their half maximal inhibitory concentration (IC50). Bac I and II at specific doses reduces the viability, proliferation, migration and invasiveness of breast cancer cell lines (Palethorpe, et al. [17]).
Bacosides comprises a family of 12 known analogs [11] called as bacopasides I–XII [12]. The alkaloids brahmine, nicotine, and herpestine have been catalogued, along with D-mannitol, apigenin, hersaponin, monnierasides I–III, cucurbitacin and plantainoside B [18,19]. The pseudojujubogenin group is composed of bacopaside I-II and bacopasaponin C (Chakravarty, et al. [19]). Few of these glycosides such as Bacoside X, bacoside A, 3-beta-Dglucosylstigmasterol and daucosterol could be good inhibitors of acetylcholinesterase (AChE) and pseudocholinesterase (BuChE). These studies suggest that the natural compounds of BM can be utilized for the development of a class of various inhibitors (Jamal, et al. [20,21]). Phytochemical and physiological effects of the different parts of a medicinal plant varies depending on the chemical composition and their relative abundances. It is well established that the phytochemical landscapes are tissue-specific and are based on the role they have in each tissue during the plant growth (Defossez. et al. [22]). Investigating the tissue-specific metabolic diversity is of prime importance to gain a comprehensive understanding of the biological functions of respective part of the plant.
Applications of metabolite and lipid profiling to plant tissues meets many challenges. Untargeted lipidomic and metabolomics render the powerful and efficient tools for the identification and quantitation of relative abundance of chemical species. These chemical species can thus be analyzed and authenticated in various tissues, species, ages, environment, and geographical origins. Limited studies exist on the metabolomics and/or lipdiomic studies in BM. Further, phytochemical landscape of lipids and metabolites in root, stem, and leaves of BM remain unclear. In this article, we compile the current state and progress of metabolomics and lipidomics analysis. The metabolome and lipidome of BM tissues were analyzed using ultra-high performance liquid chromatography high-resolution electrospray ionization mass spectrometry (UHPLC-HR-ESI-MS). Aim was to explore and identify metabolic and lipidomic signatures associated with different tissues; leaves, stem and roots of the BM. This study not only represents the first global metabolome and lipidome analysis of BM but also provides information about the identification, expression, and differential tissue specific distribution of small molecules. The present research will establish a foundation for the studies related to tissue specific plant physiology and its regulation in human for health benefits.
Reagents and Samples
Plant Materials: BM plant was obtained from the Gandhi Krishi Vigyana Kendra (GKVK) campus, Bangalore, India. The taxonomical classification was carried out at The Department of Biotechnology, BMS College of Engineering, Bangalore. Leaf, stem and root tissues were harvested from the plants using a razor blade. All the tissues were washed with raw water followed by de-ionized water and immediately frozen in liquid nitrogen and stored in -80 °C until used for the LC-HRMS analysis. Three replicates of original samples were used for metabolomics study whereas, 2 replicates for lipidomics studies. Each tissue was analyzed in triplicate for metabolites and duplicate for lipidomics.
Chemicals and Materials: All solvents were of LC-MS grade unless stated otherwise. Water, acetonitrile, and methanol were procured from Thermo Fisher. Analytical grade chloroform, butylated hydroxytoluene (BHT), from Sigma Aldrich. Splash mixture of Deuterated lipid standards was procured from Avanti polar lipids (USA). Reserpine was used for the area normalization in metabolomic experiment.
Lipid Extraction from Root, Stem, and Leaf Tissues: Lipids were extracted using modified Bligh & Dyer method (Iverson, 2001). Samples spiked with 10 μl of splash lipids standard mixture. Samples were crushed in 1.0 ml of chloroform: methanol (50:50) containing 0.01% butylated hydroxytoluene (BHT) then homogenized using Minilys personal homogenizer (Bertin Instrument, France) for 5 cycles each of 10 sec with 1-minute intermittent cooling on ice. 500 μl of chloroform was added and mixed thoroughly. Samples were centrifuged at 14000 rpm and allowed for phase separation. Lower chloroform phase was separated into new Eppendorf tube. Another aliquot of 500 μl of chloroform was added to the residue, mixed thoroughly, centrifuged, stranded for phase-separation. Chloroform layer was separated and mixed with the original chloroform extract. Solvent was evaporated in speed-vac at 4 °C. 200μl of 90% methanol was added to the residue, mixed thoroughly, and transferred into LC-MS vial.
Metabolite Extraction from Root, Stem, And Leaf Tissues: After freeze-thawing, frozen leaf, stem, and root tissues were crushed in pestle and mortal for 5 min. About 75 mg of each tissue was weighted in tubes and spiked with the 1μg/ml reserpine solution prepared in methanol. 1.0 ml of water was added to the crushed samples and homogenized thoroughly using zirconia beads in five cycles each of 10 sec with 1-minute intermittent cooling on ice. Samples were filtered through 22 μm membrane using syringe filters and 100ul of the filtrate transferred in vials for LC-MS/MS analysis. 3μl was injected on to the column.
LC-ESI-HRMS for the Lipid Profiling: Lipids were separated using a reversed-phase Acclaim C30 column (100 x 3.0 mm; 3um particle size) procured from Thermo Fisher Scientific (USA). LCHRMS data was acquired using Dionex-Q Exactive as described in following section. Mobile phase was made up of solvent A composed of 10 mM ammonium acetate and 0.1% formic acid in water whereas solvent B was 100 isopropanol. Flowrate was 300 μl/min and column was maintained at 40 °C throughout the run.
LC-ESI-HRMS for the Metabolomics: Chromatographic separation and Mass Spectrometry detection were performed using a Dionex ultra high-performance liquid chromatography (UHPLC) and high-resolution mass spectrometer (HRMS) QExactive (Thermo Finnigan, San Jose, CA, USA), equipped with an ESI source. The chromatographic separation was achieved on an Acquity BEH C18 (100 x 1.0 mm id; particle size 1.7 μm) analytical column maintained at room temperature. The optimum mobile phase was consisted of 0.1% formic acid in water as solvent A and 0.1% formic acid in acetonitrile as solvent B. The gradient elution performed as: time zero, 25% solvent B; 10 min, 60% solvent B; 15 min, 80% solvent B; 20 min, 90% solvent B; at 25 min, 90% solvent B; at 25.1 min, 25% solvent B; at 35 min, 25% solvent B. The flow rate was 1.0 mL/min, the total run time was 35 min, the column was maintained at 40°C and the auto sampler temperature was 7°C. 3 μL of sample Extract was injected for the analysis. The data acquisition was under the control of Xcalibur software (Thermo Electron Corporation, USA). The mass spectrometer was operated in positive as well as negative ion mode using polarity switching. Ions were acquired in full scan MS and data-dependent DD-MS2 mode with the scan range at 84 to 1250 m/z. Optimized spray voltage was at 5.5 kV for positive and 4.5 kV for negative mode, capillary temperature and prob-heater temperature were maintained at 300°C and 320°C respectively, sheath gas and auxiliary gas (nitrogen) pressure were maintained at 40 and 10 arbitrary units, respectively.
Nitrogen was used as collision gas at a pressure of 1.5 mTorr and stepped normalized collision energies used were 20 – 38 eV throughout the run. MS resolution for the precursor ion and MS/ MS scanning were maintained at 140000 and 35000 respectively. Dynamic exclusion of the repetitive ions was for 20 sec. Automatic gain control values for precursor and MS/MS scanning were 1e6 and 1e5 respectively.
Statistics: Lipids were identified, and peak areas of each identified lipid species was extracted using LipidSearch 4.0 software (Thermo Fisher Scientific, CA, USA) whereas metabolites were identified and annotated using the Compound Discoverer software (Thermo Fisher Scientific, CA, USA). The data sets were then imported in Partek software (Partek, USA) to visualize the data through plots and heat map. Statistical analysis such as partial least squares discriminate analysis (PLS-DA), variable importance in projection (VIP) scores were calculated using Partek.
Untargeted Lipidomics Analysis
An untargeted reversed-phase liquid chromatography-high resolution mass spectrometry (LC-HRMS) method was optimized aiming to enable the in-depth profiling, characterization, and quantitation of different classes of lipids in BM leaves, stem and roots. A hydrophobic C30 column was used to resolve the isobaric species of the lipids. A Full MS and data-dependent acquisition MS/ MS (dd-MS2) in high-resolution mode was used. LC-HRMS counter plots for BM leaves, stem and roots in positive and negative polarities are shown in (Supplementary Figure 1). The identification, relative quantitation, and calculation of estimated concentration of each identified lipid species using the spiked-in SPLASH standard mixture were carried out using Lipid Search SP2 software. All the raw files were first aligned with respect to retention time. With the directed MS-MS approach, lipid ions with very low abundance were triggered for MS-MS, yielding in-depth lipidome identification coverage over the complex plant tissue samples. The summary of lipid identification and quantitation is shown in the (Table 1). Total of 338 lipid species were identified after filtering with main ions per lipid class. The estimated concentration results were obtained for lipid species across nineteen lipid classes including AGlcSiE, BisMePA, Cer, CerG1, DAG, LPA, LPC, LPE, LPMe, MePC, MAG, PA, PC, PE, PG, PI, PS, SM and TAG using the spiked SPLASH labeled-internal standard. Number of identified lipid species and relative abundance of each lipid class in leaves, stem and roots are summarized and listed in (Table 1). Complex mixture of molecular species that show diversity in the acyl chain length and unsaturation of the fatty acyl chain at sn−1 and sn−2 positions.
Figure 1 Suplementary Figure 1: Contour plot of the LC-MS/MS separation of lipids (A-F) and metabolites (G-L). A & G are in positive mode of Bacopa leaves; B & H positive mode of stem; C & I in positive mode of roots. D & J in negative mode of leaves; E & K in negative mode of stem and F & L in negative mode of roots.

Note: *p< 0.05; **p< 0.01; ***p< 0.001
Note: 10 μL of the splash was spiked and final reconstitution volume was 200 μL (20 times dilution).
A total of 13 differential lipid classes were identified as significantly altered between leaves, stem and roots. Most of the lipid species are highly abundant in leaves compared to stem and roots. Highest number of lipid species from single class were TAG followed by PE. PIs were the highest abundant species in leaves and stem whereas PGs were the most abundant species in roots (Table 2). The largest difference was a decrease in fatty acid carbon in phospholipids in roots compared to leaves and stem. Phospholipids containing the 38 carbons in total in both fatty acids were significantly lower in roots compared to leaf and stem. In roots, fatty acids having 36 carbons are predominant. AGlcSiE (16:1) was the most abundant in leaves followed by stem and roots. AGlcSiE (18:3) was the highest level among seven lipid species of AGlcSiE in stem and roots. In phospholipids, the major molecular species of PC, PE, PI were PE (40:5). BM leaves and stem contain proportionally more PI, PE and PC than roots. In addition, fatty acid moieties in phospholipids in leaves and stem are longer and more unsaturated than roots and this phenomenon is even stronger in the leaves than in the stem (Figures 1F & 1G). Similar trend was observed in neutral lipids (TAG and MAG). Roots have higher PG and DAG than other two tissues. The fatty acid in sphingolipids in roots had increased average length than the stem and leaves (Figure 1H). Whereas sphingolipids in leaves and stem had higher degree of unsaturation than in roots (Figure 1I).
Figure 2 Lipid class profiles vary in different tissues A. Lipid class abundance is presented nanogram per milligram of the tissue; Fatty acid chain length and unsaturation varies in different tissues. B. (B–I); The distributions of combined fatty acid chain length (left panels) and unsaturation (right panels) in (B & C) TAG (D & E) DAG (F & G) phospholipids (H & I) sphingolipids from different tissues. The abundance of each is presented as a percent relative to all lipids in its category. All the values represented are the mean of the two biological replicates.

Figure 3 Volcano plot comparison of relative abundance of metabolites (Top panel) and Lipids (Lower panel) between leaves and roots (A & D); leaves and stems (B & E); and stem and roots (C & F); Green circles: down-regulated; Gray circle: unchanged; Red circles: up-regulated.

Phosphatidylcholine (PC) species such as PC (16:0/18:1); PC(16:0/18:3); PC(20:0/14:1) levels were highest in leaves than roots and stem whereas PC(16:0/18:2) highest in stem and roots compared to leaves. Total PC concentration was 32-fold and 4-fold higher in leaves compared to roots and stem respectively. PC levels were >7-fold lower in roots compared to stem. Phosphatidylethanol such as PE (40:5) in leaves; PE(16:0/18:2) in stem; PE(32:1) in roots were the highest abundant. Total PE levels were 3.2-fold and 3.9-fold higher in leaves compared to stem and roots respectively whereas PE levels in stem and roots were similar. Phosphatidylinositol (PI) levels in leaves were 0.7-fold lower compared to stem whereas 12- fold higher than roots. Stem PI levels were 18-fold higher compared to roots. Roots showed highest levels of phosphatidylglycerol (PG) comprising at 5-fold and 2.4-fold compared to leaves and stem. Out of nineteen PG species, PG(18:1p/10:1) comprises 73.4%, 84% and 92% of total PG concentration in leaves, stem and roots respectively. Triglyceride (TAG) levels were 1.60-fold higher in leaves and roots compared to stem. Levels of TAG(16:0/18:3/18:3) were observed highest in leaves, whereas TAG(4:0/10:4/12:3) higher in stem and TAG(4:0/10:4/11:1) higher in roots. Diglycerides (DAG) levels were highest in roots at 2.7-fold than leaves. DAG(18:0/18:0) levels were higher in roots whereas DAG(18:0/16:0) levels were higher in stem and leaves compared to roots. Volcano plots were used to study the phytochemical differences of lipids between the leaf, stem and root tissues. Lower pane of (Figure 2) represents the differences in lipid levels between leaf vs root (D); leaf vs stem (E) and stem vs root (F).
Un-Targeted Metabolomics Analysis
We putatively identified 93 small biomolecules other than the listed lipids using Compound Discoverer (Thermo Finnegan, USA). (Supplementary Table 2) represents the detailed identification of all these biomolecules. Compound characterization was validated using the highly resolved precursor ions with the mass tolerance (Δm) ≤ 5 ppm, reported fragmentation pattern, and limiting mass error of the fragments at Δm ≤ 5 ppm. Compounds were annotated using mzCloud, mzLogic, Chemsspider search and Metabolika pathways mapping (Workflow available with Compound Discoverer software). Further, compound identification was supported by a suite of tools such as KEGG, PlantCyc, BioCyc, and Metabolika biological pathway databases available with the software. The relative abundance of each metabolite between leaves, stem and roots was compared in order to identify the compounds that are differentially accumulated in these three tissues. PCA, volcano plots and hierarchical clustering of these metabolites are the widely used techniques to present the data effectively that helps to identify the differences among the groups. Peak areas of all detected and annotated metabolites are mentioned in (Table 1). Based on the quantitative metabolic profiles of the three tissues, principal component analysis (PCA), volcano plots and heatmap hierarchical clustering could clearly distinguished leaf samples from stem and roots; stem samples from leaf and roots suggesting that although these tissues contain the approximately the similar metabolite profile, their relative abundances are varying considerably among tissues.
Levels of bacopaside A oxy-p-2glc-malonylpentoside are highest is leaves and lowest is roots, whereas butylparaben levels in roots higher than leaves as well as stem. Top ten metabolites with the highest peak area found in each tissue are presented in (Figure 3).
Figure 4 A) Comparison of ten most abundant metabolites observed in Leaves, stem and roots; Ten most abundant metabolites in B) Leaves, C) Stem, D) Roots.

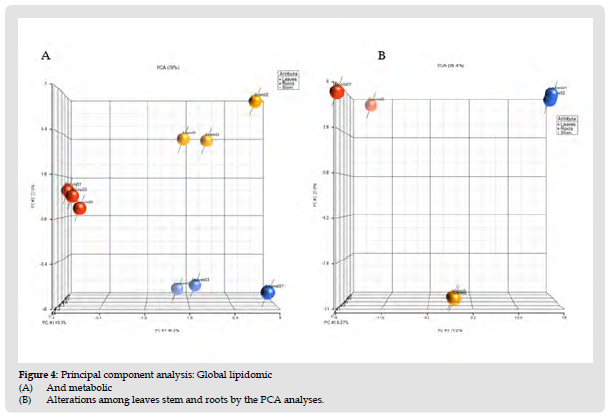
The PCA model shows a significant grouping between leaves, stem and roots samples with no points overlapped, illustrating the remarkable difference between three tissues with respect to their phytochemical content. The significant variation in metabolite levels between leaves, stem and roots observed in the present study suggests that the medicinal importance and physiological effects of each tissue may be different. To study phytochemical landscape of each tissue, we compared the results using volcano plots in (Figure 2). Upper pane of (Figure 2) represents the metabolite comparison between leaf vs root (A); leaf vs stem (B) and stem vs root (C). The significant metabolites were selected based on the fold changes ≥ 2 or fold change ≤ 0.5. In total 32 metabolites accumulated differentially between the leaves and stem (Figure 4) and Table 3. Levels of twenty metabolites were higher in leaves compared to stem whereas low abundance of 12 metabolites in leaves were observed compared to stem. Bacopaside A oxy-p-2glcmalonylpentoside levels in leaves were significantly higher than the combined peak areas of the metabolite in roots and stem. Levels of phytochemicals such as bacopaside A3, bacopaside I, bacopaside II which are responsible for the memory enhancing effects and also known as the markers of BM are highest in leaves compared to stem and roots. Levels of these metabolites are higher in stem compared to roots. These saponins are at high levels in leaves compared to stem and roots indicating that leaves may have more strength to improve the memory. From the heatmap, three main groups of metabolites were obtained in which some metabolites were highly accumulated in the stems and roots and some metabolites at higher levels in stem compared to leaves and roots.
Figure 5 Principal component analysis: Global lipidomic (A) And metabolic (B) Alterations among leaves stem and roots by the PCA analyses.
Heatmap Analysis
The heatmaps based on the identified endogenous metabolites and lipids were prepared to show the overall variation in distribution of metabolites and lipids in all three BM tissues. The heat maps were constructed using a hierarchical clustering algorithm based on normalized peak areas of metabolites. The red and green colors in both the heatmaps represent higher and lower relative levels than average value respectively. Heatmap distribution of metabolites and lipids is clearly distinguished, suggesting the significantly different levels of metabolites and lipids among tissues. The left panel A of the (Figure 5) shows the relationships between the lipid profiles of roots, leaves and stem. The major difference was found in the levels of bacopaside I and II, leteolin, oxy-bacopaside I, 3-Hydroxymandelic acid and bacopasaponin F which are highest in leaves compared to stem and roots. Several metabolites such as dextromethorphan, 1,2,4-butanetriol, encecalin, desmethylselegiline, 9,12-octadecadienal, Monoolein, glycidyl oleate, Leucomalachite green, j-ara-glc- are higher in roots compared to other two tissues whereas, (+/-)12(13)-DiHOME, Acetylcholine, and proline levels are higher in BM stem. These results indicates that leaves, stem and roots display different phytochemical profiles which may enables them to respond differently to the environmental factors as well as may have different medicinal properties as well as efficacy.
Figure 6 Hierarchical clustering analysis of differently expressed lipids (A) And metabolites (B) Found in leaves (left panel), stem (middle panel) and roots (right panel). Each coloured bar within a column represents a molecular species (Lipid and/or metabolite) in the indicated plant part. The colour of each bar in Figure 4A represents the levels of corresponding lipid species (ng/mg wet weight) whereas the colour of each bar in Figure 4B represents normalized peak area of each metabolite.

Heatmap of lipid profile in roots, stem and leaves suggests that most of the lipid levels across all the classes except PG and PIs are highest is leaves compared to other tissues. PG levels are more than 4-fold higher in roots compared to leaves and >2-fold compared to stem. PG levels are >2-fold higher in stem compared to leaves. PI levels in stem are >1.4-fold higher compared to leaves and significantly higher (>18-fold) compared to roots.
Versatile analytical approaches with greater efficiency and accuracy through high-throughput screening are required for deep convolution and wide coverage of small molecules in plant tissues. In the current work, we demonstrated the development and applications a highly sensitive and accurate UHPLC-HRMS for the identification of natural products viz., metabolites and lipids in BM. UHPLC-Orbitrap MS platform incorporated with Full MS/dd- MS2 enabled presented a simultaneously targeted and untargeted approach for metabolite annotation approach which helped not only to establish a comprehensive phytochemical landscape of BM but also showed a contrasting diversity of bioactive metabolites in different parts of a BM plant. This phytochemical metabolite landscape associated with the BM plant is powerful information to support in the development of more effective and specific applications of BM in improving memory and treatment of neurological disorders.
Authors acknowledge the support from Mass Spectrometry core, NCBS, Bnagalore for LC-MS/MS analysis.


