Abstract
The E26 transformation specific (ETS) transcription factor family contains approximately 28 members in humans. EHF (ETS homology factor) is a transcription factor that is generally expressed in epithelial tissue and has been shown to play a role in a variety of human carcinomas. Research from a number of studies implicate EHF in gastric cancer, ovarian cancer, prostate cancer, colorectal cancer, and pancreatic cancer. Interestingly, EHF can act both as a tumor activator and tumor suppressor in different human cancers. As an activator, EHF overexpression activates cellular differentiation and proliferation mechanisms. As a suppressor, EHF promotes the expression of several miRNAs and blocks key oncogenic transformation pathways. The current bioinformatics report provides strong evidence of the potential therapeutic value of EHF. Data mining procedures involving human primary prostate epithelial cells (HPEC) and DU145 prostate cancer cells revealed that the EHF transcription factor was significantly downregulated in DU145 human prostate cancer cells compared to HPEC cells suggesting that it plays a role in tumor suppression in DU145 cells.
In fact, Ingenuity Pathway Analysis (IPA) revealed that EHF showed the highest level of down-regulation (expression fold change) than any gene in the dataset. Upstream regulator analysis demonstrated that EHF leads to the predicted activation of CCBE1 and the predicted inhibition of FOXN1 and other proteins believed to produce anti-tumor effects in prostate cancer and other human carcinomas. Data showed EHF expression leads to inhibition of the growth of tumors and may lead to the inhibition of cell movement of tumor cell lines and migration of tumor cell lines although the analysis did not yield strong association. The purpose of this article is to present biological networks associated with EHF generated using the comparative microarray dataset. Clinical strategies that focus on EHF may prove to be effective weapons in the armamentarium of therapies to treat prostate cancer and potentially other human carcinomas.
Keywords: Transcription Factor; Oncogene; Bioinformatics; Prostate Cancer; Microarray
Abbreviations: IPA: Ingenuity Pathway Analysis; HPEC: Human Primary Prostate Epithelial Cells; EHF: ETS Homology Factor; ETS: E26 transformation specific; TGF-β: Transforming Growth Factor Beta; HPrEC: Human Primary Prostate Epithelial Cells
Introduction
The ETS (E26 transformation-specific or erythroblast transformation specific) homologous factor, EHF, is an emerging epithelium-specific transcription factor that plays a role in many human cancers [1]. All ETS family members contain an ETS DNA-binding domain that allows the helix-turn-helix structure to recognize a specific eukaryotic DNA sequence called the ETSbinding site. The ETS family contains 12 subfamilies (e.g., ELF, ELG, ERG, ERF, ESE, ETS, PDEF, PEA3, ER71, SPI, TCF, TEL). Specifically, EHF contains approximately 300 amino acids and is a member of the subfamily ESE (epithelial-specific expression). EHF is located on human chromosome 11 (11p13) [2,3]. ETS proteins regulate many biological functions such as cellular proliferation, differentiation, cell and tissue development, and tumorigenesis [4]. ESE family members also contain an N-terminal pointed (PNT) domain that is purported to participate in various biomolecular interactions [5,6]. Like most transcription factors, EHF regulates the transcription rate of many genes in different cell types. ETS family members have previously been shown to disrupt normal cell proliferation processes and promote oncogenesis through the use of fusion proteins.
One of the most studied fusion proteins shown to promote abnormal cell growth and metastasis in humans is the TEL-JAK fusion protein [7]. Chromosomal translocation mechanisms and human pathophysiological outcomes of the TEL-JAK2 model system were actively studied in the latter part of the 20th century and early part of the 21st century in human leukemia [7-9]. These researchers found that the TEL-JAK fusion protein resulted in a constitutively active tyrosine kinase which subsequently activated many signaling pathways (e.g., JAK-STAT, p38 signaling pathway) that regulate proliferation and other physiological processes that mediate oncogenesis and tumor progression. The purpose of the current investigation was to utilize bioinformatics analysis to create network visualizations that highlight the paramount role of EHF in prostate cancer progression.
EHF as a Tumor Activator
Induction of tumorigenesis was investigated in cells in which EHF is abnormally expressed. Ovarian cancer researchers found that overexpression of EHF in ovarian cancer cells led to adverse clinical outcomes and truncated survival periods [10]. EHF knockdown studies provided evidence that EHF reduction in ovarian cancer cells could attenuate cell growth, metastatic potential, block essential cell cycle proteins, and accelerate apoptosis. The ERK and AKT signaling pathways contribute to anti-tumor biological mechanisms. Colorectal cancer investigators demonstrated that higher EHF levels in colorectal cancer tissues correlated with reduced patient survival, enhanced cellular invasiveness, and uncontrolled cell expansion mediated via the TGF-β (Transforming growth factor beta) signaling pathway [11]. Both disrupting EHF binding and inhibiting the TGF-β pathway may positively affect ovarian cancer cells. According to a previous study in gastric cancer, EHF was significantly elevated compared to non-gastric cancer control patients [1].
Researchers utilized various experimental techniques (qRTPCR, IHC, FISH) to show that gene amplification mechanisms caused increased EHF expression. Additionally, knocking down EHF expression using siRNAs specific to EHF was shown to affect gastric cancer cell density and cellular migration potential negatively. Further analysis revealed that EHF was responsible for activating or modulating members of the human epidermal growth factor receptor (HER) family. This experiment provided therapeutic proof-of-concept that direct interference with EHF gene expression in aggressive gastric cancer can lead to beneficial clinical outcomes. MicroRNA‐206 (miR-206) has been identified to inhibit the growth of gastric cancer tissues and cell lines [12]. miR-206 expression has been shown to disrupt oncogenic mechanisms by targeting several signaling pathways including the c-Met and mitogen activating protein kinase [13,14] and diminish the effects of cyclinD2, a protein that plays a role in the G1/S cell cycle transition [15,16]. Using the HER2-positive gastric cancer cell line, NCI-N87, researchers demonstrated that miR-206 overexpression effectively inhibited EHF protein production, leading to the suppression of cancer-initiating cell functional traits that made NCI-N87 cells more susceptible to 5-fluorouracil [17]. The evidence-based efficacy of targeting EHF to reduce tumor size suggests that EHF’s therapeutic probative value is justifiable.
EHF as a Tumor Suppressor:
Surprisingly, EHF also shows suppressive actions of prostate cancer cells and attenuates the prostate cancer stem-like compartment’s growth Albino et al. [18] demonstrated that EHF regulated IL-6 expression and blocked STAT3 activation and abnormal prostatic transformation. CHiP binding studies confirmed the ETS binding site in the IL-6 gene promoter region. Blocking IL-6 via tyrosine kinase inhibitor peptide was an effective strategy to target the JAK-STAT pathway [19]. Researchers explored the clinical outcome when EHF expression was reduced. They found that with prostate cancer, EHF regulated the production of Lin28A, Lin28B, and let-7 microRNAs [20]. The increase of Lin28A/Lin28B and downregulation of let-7 microRNAs conferred tumor-like properties on the cells under study. While different methods were utilized, the current study provides bioinformatics data in DU145 prostate cancer cells that portray EHF as a tumor suppressor. In DU145 cells, EHF is downregulated when compared to normal prostate cells. A rationale that explains why EHF acts as a tumor suppressor in prostate cancer and acts as an activator in many other human carcinomas is unclear and certainly warrants further investigation.
However, it suggests that tissue-specific gene expression mechanisms may coordinate different functional outcomes in different parts of the human body Cangemi et al. [21] showed that EHF expression attenuation was caused by methylation events in clinical prostate cancer samples. The reintroduction of EHF in prostate cells promoted proapoptotic molecules such as procaspase-3 by binding to promoter regions associated with caspase-3. In addition to prostate cancer cells, EHF also functions as a tumor suppressor protein in pancreatic cancer cells [22,23]. In pancreatic ductal adenocarcinomas EHF was shown to block tumor invasion transformation by initiating the expression of e-cadherin, a molecule important for mediating cellular adhesion and suppressing metastasis. The loss of EHF in pancreatic cancer cells produces metastatic phenotypes and lugubrious prognoses for patients [22]. Recently, Liu et al. [23] showed the efficacy of utilizing EHF to portend the potential functionality of the monoclonal antibody, anti-PD1, to treat pancreatic ductal adenocarcinoma. PD1 is the programmed cell death receptor that when active in tumor cells leads to a weakened immune system that is ineffective at fighting cancer [24].
When EHF was highly expressed in pancreatic cancer cells anti-PD1 therapy was more effective at reducing tumor size in cancer mouse models, suggesting that EHF has the capacity to reverse immunosuppressive activity in cancer cells. EHF was also suspected to be involved in suppressing traits associated with esophageal squamous cell carcinoma. Moreover, aberrant subcellular localization of EHF may predict the biological outcome of EHF in esophageal squamous cells [25]. EHF was found to be located in the nuclear region of healthy esophageal squamous cells following cell biology experiments and found to be located in the cytoplasm of esophageal cancer cells suggesting that nuclear EHF is a requirement for regulating cancer progression in these cells. Increased expression of EHF through plasmid-based technology restored the nuclear localization of EHF which showed a reduction in tumor-like properties [25].
Materials and Methods
Cell Culture
Non-cancerous HPrEC (human primary prostate epithelial cells) and the human prostate cancer cell line, DU145, were purchased from ATCC. Cells were cultured as previously described and viewed under an inverted compound light microscope. DU145 cells were harvested at 90% confluence. Trypan blue exclusion techniques were utilized to determine percent viability and for enumeration protocols [26].
Microarray Sample Preparation
Purified cellular RNA was collected from HPrECs and DU145 cells using an RNA extraction kit (RNeasy Plus Mini Kit) from Qiagen. Prostate cell RNA samples were then submitted to the Duke University Microarray Facility for microarray processing that included RNA quality control assessments, labeling, hybridization, and scanning. The experimental design included appropriate biological replicates to evaluate variability in the dataset. Different biological replicates produced similar results according to analytical measurements.
Bioinformatics Analysis
Using previously published methods [26], IPA was utilized to analyze the microarray dataset with respect to EHF.
Results
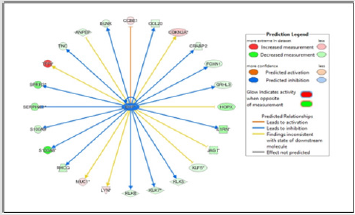
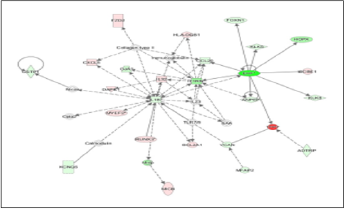
In Figure 1 upstream regulator analysis was performed using IPA to predict upstream molecules most likely affected by EHF, according to statistical analysis. The figure legend highlights important characteristics such as predicted measurement amount, activity, and relationships. Putatively, EHF is predicted to increase expression of CCBE1. EHF is predicted to decrease the expression of FOXN1, SPRR3, BLNK, and HOPX. The data reveals that EHF may play a largely inhibitory function (blue lines) of key molecules known to play a role in oncogenesis from a variety of cell types. Figure 2 shows a major molecular network associated with EHF in prostate cancer cells. This depiction shows an almost equal number of upregulated molecules as downregulated molecules but does show other molecules that may alter EHF expression and presumably function in DU145 cells. The top network in the dataset contains various molecules that contribute to tumorigenesis in the prostate and other tissue, such as ANPEP, CCL20, KLK5, IL1RN, and KLK8. Identification of FOXN1 and HOPX in this analytical framework is consistent with findings in Figure 1 (upstream regulator analysis). The network analysis depicts many indirect molecular relationships (broken line). Figure 3 shows the result of bioinformatics analysis that correlate biological functions with molecular networks associated with EHF.
Data shows how molecules depicted in previous models are statistically linked with functional processes purported to influence tumor development and poor cancer survival outcomes such as the growth of tumor (Figure 3), cell movement of tumor cell lines (Figure 3), and migration of tumor cell lines (Figure 3). EHF is associated with several oncogenic and metastatic processes; however, the top networks are shown in Figure 3. In the growth of tumor analysis (Figure 3), several molecules with decreased measurement, including Mmp, GJA1, IL-1B, VCAN, IL1RN, and ANPEP with EHF showing the highest reduction compared to control cells (e.g., highest color intensity). Network molecules associated with cell movement of tumor cell lines consist of similar molecules observed in Figure 2. CXCL5, IL-32, and RUNX3 show a decreased measurement in each functional molecular analysis. Interestingly, while IL-32 shows increased production in all three figures, IL-32 is predicted to inhibit the tumor’s growth indirectly and is expected to indirectly activate both cell movement of tumor cell lines and migration of tumor cell lines. According to the data, EHF only showed an indirect relationship with the biological processes under study, suggesting that functional assays regarding EHF to examine in silico relationships are warranted.
Discussion
EHF (ESE3) is a key member of the ETS transcription factor family and is largely defined by the ability to regulate differentiation and cell proliferation processes in epithelial cells [27]. EHF is associated with many human cancers and functions as either a tumor activator or tumor suppressor. There are mixed results when examining EHF in the context of tumorigenesis. There are several papers that suggest that EHF functions as a tumor activator and also research studies that provide evidence that EHF is a tumor suppressor. The current study is more aligned with the latter scenario. Computational investigations showed that the transcription factor, EHF, was significantly downregulated in DU145 cells compared to normal primary prostate epithelial cells. Bioinformatics analysis indicates that EHF may exert its function by regulating gene expression of critical genes that control and activate tumor progression (e.g., TFPI) and by regulating gene expression that assist in the suppression of human carcinomas (e.g., FOXN1). Tissue factor pathway inhibitor (TFPI) was significantly upregulated in a human prostate cancer cell line suggesting its role as a tumor activator.
TFPI expression has been shown to enhance tumor progression in human breast and colon cancer [28]. Lwaleed, Lam, Lasebai, and Cooper [29] also demonstrated that TFPI was upregulated in prostate cancer cells compared to normal prostate epithelial cells. Forkhead box N1 (FOXN1) acts as a tumor suppressor protein in non-small cell lung cancer [30] and appears to function in a similar manner in prostate cancer since it was shown that FOXN1 is downregulated in the dataset Ji et al. [30] found that reduction of FOXN1 in respiration cell line models produced tumorigenic phenotypes while increased expression of FOXN1 led to attenuation of traits associated with invasive cancerous cells such as unregulated cellular migration to ectopic locations. There are very few studies however, implicating FOXN1 in specific anti-cancer mechanisms in prostate cancer cell lines. In general, tumor suppressor proteins like EHF decrease the transformation of normal cells to cancerous cells. However, an unusual protein, p16(Ink4a), that mediates control of cell cycle events has been shown to act as a tumor suppressor and as a tumor activator when overexpressed [31].
Several members of the SOCS protein family have been explored as potential therapeutic solutions to treat human carcinomas including prostate cancer. SOCS proteins are generally regarded as tumor suppressor proteins that use a negative feedback loop to limit cytokine signaling events through blocking autophosphorylation events [32]. Previous observations have demonstrated very positive in vitro outcomes using tumor suppressor protein mimetics and overexpressed tumor suppressor proteins in prostate cancer cell lines. A small 12-amino acid SOCS1 peptide mimetic [33] was shown to ameliorate cell proliferation and cell cycle progression in DU145 and LNCaP prostate cancer cells by targeting STAT3 and IL-6 signaling [34]. SOCS6 is a member of the suppressors of cytokine signaling proteins that function as inducible negative regulators of signal transduction pathways that control differentiation, proliferation, and key cellular physiologic processes. Yuan et al. [35] performed comparative SOCS6 expression analysis in prostate cancer tissue and healthy prostate tissue using immunohistochemistry.
SOCS6 was shown to be more highly expressed in non-cancerous prostate tissues compared to cancer tissue. Further, when SOCS6 was overexpressed in prostate cancer cells, angiogenesis and tumor development was repressed and cellular apoptosis was increased. Microarray datasets are intriguing and beneficial tools to explore gene expression profiles in cells, tissues, and diseases and represents opportunities to identify biomarkers and regulatory genes important in transformation events [36,37]. When viewing EHF as a tumor activator, subsequent bioassays involving smallinterfering RNA (si-RNA) will be employed to adduce biological function. As was briefly mentioned earlier in the report, increased in vivo, in vitro, and in silico investigations should be conducted to learn more about potential miRNAs that are capable of blocking EHF directly or blocking downstream genes or promoter elements associated with EHF as a means to control tumorigenesis. Conversely, when viewing EHF as a tumor suppressor, EHF overexpression studies may provide additional evidence of EHF as a beneficial transcription factor with therapeutic value. While data presented in this article and subsequent conclusions drawn are based on statistical procedures and not actual functional assays, the data provided herein are useful to subsequent research investigations by revealing molecular targets of EHF and other potentially more reliable therapeutic targets for clinical intervention.
Acknowledgements
This work was supported by a grant funded by the National Science Foundation (HRD-1533536).
References
- Shi J, Qu Y, Li X, Sui F, Yao D, et al. (2016) Increased expression of EHF via gene amplification contributes to the activation of HER family signaling and associates with poor survival in gastric cancer. Cell Death & Disease 7(10): 1-14.
- Bochert M, Kleinbaum, Sun L, Burton F (1998) Molecular cloning and expression of Ehf, a new member of the ets transcription factor/oncoprotein gene family. Biochemical and Biophysical Research Communications 246(1): 176-181.
- Kleinbaum L, Duggan C, Ferreira E, Coffey G, Butticè G, et al. (1999) Human chromosomal localization, tissue/tumor expression, and regulatory function of the ets family gene EHF. Biochemical and Biophysical Research Communications 264(1): 119-126.
- Nicholas T, Strittmatter B, Hollenhorst P (2019) Oncogenic ETS factors in prostate cancer. Advances in Experimental Medicine and Biology 1210: 409-436.
- Luk I, Reehorst C, Mariadason J (2018) ELF3, ELF5, EHF and SPDEF transcription factors in tissue homeostasis and cancer. Molecules 23(9): 1-25.
- Laudet V, Hänni C, Stéhelin D, Duterque Coquillaud M (1999) Molecular phylogeny of the ETS gene family. Oncogene 18(6): 1351-1359.
- Lacronique V, Boureux A, Valle V, Poirel H, Quang C, et al. (1997) A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science 278(5341): 1309-1312.
- Schwaller J, Frantsve J, Aster J, Williams I, Tomasson M, et al. (1998) Transformation of hematopoietic cell lines to growth-factor independence and induction of a fatal myelo- and lymphoproliferative disease in mice by retrovirally transduced TEL/JAK2 fusion genes. EMBO Journal 17(18): 5321-5333.
- Ho J, Nguyen M, Dierov J, Badger K, Beattie B, et al. (2002) TEL-JAK2 constitutively activates the extracellular signal-regulated kinase (ERK), stress-activated protein/Jun kinase (SAPK/JNK), and p38 signaling pathways. Blood 100(4): 1438-1448.
- Cheng Z, Guo J, Chen L, Luo N, Yang W, et al. (2016) Knockdown of EHF inhibited the proliferation, invasion and tumorigenesis of ovarian cancer cells. Molecular Carcinogenesis 55(6): 1048-1059.
- Wang L, Ai M, Nie M, Zhao L, Deng G, et al. (2020) EHF promotes colorectal carcinoma progression by activating TGF-β1 transcription and canonical TGF-β Cancer Science 111(7): 2310-2324.
- Ren J, Huang H, Gong Y, Yue S, Tang L, et al. (2014) MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell & Bioscience 4: 1-10.
- Zheng Z, Yan D, Chen X, Huang H, Chen K, et al. (2015) MicroRNA-206: Effective inhibition of gastric cancer progression through the c-Met pathway. Public Library of Science One 10(7): 1-11.
- Chen Z, Gao Y, Hou R, Ding D, Song D, et al. (2019) MicroRNA-206 facilitates gastric cancer cell apoptosis and suppresses cisplatin resistance by targeting MAPK2 signaling pathway. European Review for Medical and Pharmacological Sciences 23(1): 171-180.
- Zhang L, Liu X, Jin H, Guo X, Xia L, et al. (2013) miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Letters 332(1): 94-101.
- Shi H, Han J, Yue S, Zhang T, Zhu W, et al (2015) Prognostic significance of combined microRNA-206 and cyclinD2 in gastric cancer patients after curative surgery: A retrospective cohort study. Biomedicine & Pharmacotherapy 71: 210-215.
- Gong Y, Ren G, Liu B, Li F, Zhao H, et al. (2017) miR‑206 inhibits cancer initiating cells by targeting EHF in gastric cancer. Oncology Reports 38(3): 1688-1694.
- Albino D, Civenni G, Rossi S, Mitra A, Catapano C, et al. (2016) The ETS factor ESE3/EHF represses IL-6 preventing STAT3 activation and expansion of the prostate cancer stem-like compartment. Oncotarget 7(47): 76756-76768.
- Flowers L, Subramaniam P, Johnson H (2005) A SOCS-1 peptide mimetic inhibits both constitutive and IL-6 induced activation of STAT3 in prostate cancer cells. Oncogene 24(12): 2114-2120.
- Albino D, Civenni G, Dallavalle C, Roos M, Jahns H, et al. (2016) Activation of the Lin28/let-7 axis by loss of ESE3/EHF promotes a tumorigenic and stem-like phenotype in prostate cancer. Cancer Research 76(12): 3629-3643.
- Cangemi R, Mensah A, Albertini V, Jain A, Mello Grand M, et al. (2008) Reduced expression and tumor suppressor function of the ETS transcription factor ESE-3 in prostate cancer. Oncogene 27(20): 2877-2885.
- Zhao T, Jiang W, Wang X, Wang H, Zheng C, et al. (2017) ESE3 inhibits pancreatic cancer metastasis by upregulating E-cadherin. Cancer Research 77(4): 874-885.
- Liu J, Jiang W, Zhao K, Wang H, Zhou T, et al. (2019) Tumoral EHF predicts the efficacy of anti-PD1 therapy in pancreatic ductal adenocarcinoma. Journal of Experimental Medicine 216(3): 656-673.
- Sun C, Mezzadra R, Schumacher T (2018) Regulation and function of the PD-L1 checkpoint. Immunity 48(3): 434-452.
- Wang L, Xing J, Cheng R, Shao Y, Li P, et al. (2015) Abnormal localization and tumor suppressor function of epithelial tissue-specific transcription factor ESE3 in esophageal squamous cell carcinoma. Public Library of Science One 10(5): 1-14.
- Flowers L (2020) Comparison of the gene expression profiles of human primary prostate epithelial cells and DU145 prostate cancer cells. Biomedical Journal of Scientific & Technical Research 28: 21690-21696.
- Feldman R, Sementchenko V, Watson D (2003) The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Research 23(3): 2125-2131.
- Sierko E, Wojtukiewicz M, Zimnoch L, Kisiel W (2010) Expression of tissue factor pathway inhibitor (TFPI) in human breast and colon cancer tissue. Thrombosis and Haemostasis 103(1): 198-204.
- Lwaleed B, Lam L, Lasebai M, Cooper A (2013) Expression of tissue factor and tissue factor pathway inhibitor in microparticles and subcellular fractions of normal and malignant prostate cell lines. Blood Coagulation & Fibrinolysis 24(3): 339-343.
- Ji X, Ji Y, Wang W, Xu X (2018) Forkhead box N1 inhibits the progression of non-small cell lung cancer and serves as a tumor suppressor. Oncology Letters 15(5): 7221-7230.
- Romagosa C, Simonetti S, López Vicente L, Mazo A, Lleonart M, et al. (2011) p16(Ink4a) overexpression in cancer: A tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30(18): 2087-2097.\
- Washington E, Flowers L (2018) SOCS1 and SOCS3: Potential chemotherapeutic targets for human prostate cancer. International Journal of Biology, Pharmacy and Allied Sciences 7: 261-270.
- Flowers L, Johnson H, Mujtaba M, Ellis M, Haider S, et al. (2004) Characterization of a peptide inhibitor of JAK2 that mimics SOCS-1 function. Journal of Immunology 172: 7510-7518.
- Subramaniam P, Flowers L, Haider S, Johnson H (2004) Signal transduction mechanism of a peptide mimetic of interferon-gamma. Biochemistry 43(18): 5445-5454.
- Yuan D, Wang W, Su J, Zhang Y, Luan B, et al. (2018) SOCS6 functions as a tumor suppressor by inducing apoptosis and inhibiting angiogenesis in human prostate cancer. Current Cancer Drug Targets 18(9): 894-904.
- Hamilton S, White E, Washington E, Flowers L (2016) Utilization of microarray analysis to determine therapeutic targets in human cancers. International Journal of Biosciences 8: 95-105.
- Flowers L (2019) Bioinformatics analysis to explore ischemic stroke. International Journal of Biology, Pharmacy and Allied Sciences 8: 1290-1299.

 Research Article
Research Article