Abstract
Background: Changes in gene expression are increasingly used to evaluate the effects of exposure to environmental agents. Housekeeping genes (Hk) are essential in these analyzes as internal controls to normalize expression levels assessed in realtime PCR (RT-PCR). Ideal Hk genes (i) are constitutively expressed; (ii) do not respond to external stimuli and (iii) show small or no variation between samples or from one assay to another. Previous studies indicate that some commonly used Hk genes, including Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) and beta-actin, have differential expression in various cell lines. The objective of this study is, first, to identify and validate the most appropriate housekeeping genes for RNA expression analysis of human primary Peripheral Blood Mononuclear Cells (PBMCs) in response to antigen like stimulation and then to verify if selected gene can be used for isolated immune population as regulatory T cells (Treg).
Results: Using reverse transcriptase RT-PCR protocol, we show that following activation only B2M remain unchanged in PBMCs, which is also true regarding different donors in Tregs. UbC shows non- significant difference between donors in PBMC and isolated Treg. GAPDH and HPRT expression slightly modulate regarding PBMC donor, this small variation is also observed in Treg. In contrast, 18S and ATCB show significant variation in PBMC gene expression.
Conclusion: Although our results suggest that the relevance of Hk genes should be determined for each experimental condition, B2M appear to be excellent candidate as internal controls followed by UbC.
Keywords:Housekeeping Genes; RT-PCR; Human Primary PBMC; RNA Extraction; Glycolytic Enzyme
Abbreviations: GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase; PBMCs: Peripheral Blood Mononuclear Cells; HK: Housekeeping
Introduction
Housekeeping (Hk) genes are essential for quantitative gene
expression techniques interpretation. By definition, a Hk gene is
constitutively expressed in a stable manner in all nucleated cell
types, as these genes expression are essential for cell survival and
functions [1]. The Hk genes use allows to manage potential errors
committed during RNA extraction or reverse transcription, as it is
used as reference to normalize total RNA expression [2]. Indeed, Hk
gene expression should be constant in any cell type or activation state. If not, the normalization by a variable Hk gene will lead to
potential errors in gene expression quantification. Moreover,
if there is a variation in Hk gene expression, small differences in
genes of interest expression will not be detected [2]. However,
there is no single gene known with a constant expression level in
all cell type or in all activation state [3]. According to the MIQE
(Minimum Information for publication of Quantitative Real-Time
PCR Experiments) guidelines, the normalization should be made
on more than one reference gene [4].
Many studies show that some Hk genes previously recognized
as a gold standard are not as stable as thought and that Hk gene
transcription can vary significantly between different donors,
activation states, development stages, cell origins or experimental
conditions [3]. There is no reports of stable and robust primary
human PBMC Hk gene in literature . In this study, we have selected
gene described in literature to be good Hk genes in different
immune cell lines to test them in PBMCs. Indeed, as they are
described to be used in immune cell line or in isolated immune
cell population, they are most likely to be stable in PBMC. The
six genes carefully chosen are (i) β-2Microglobulin (B2M), a
component of Major Histocompatibility Complex One (MHCI), (ii)
Glycerolaldehydphosphate Dehydrogenase (GAPDH), a glycolytic
enzyme, (iii) Hypoxanthine-Guanine Phosphoribosyl Transferase
(HPRT), involved in the metabolic salvation of purines, (iv) 18S
ribosomal RNA (18S) which is part of ribosomal small subunit
, (v) Ubiquitin C (UbC) involved in protein catabolism and finally
(vi) β-actin (ACTB) a component of the microfilament part of the
cytoskeleton [1,5-8]. Our aim is to identify at least one reliable Hk
gene to use it as reliable reference for RT-qPCR on human primary
Peripheral Blood Mononuclear Cells (PBMC) and to go further to
confirm the selected genes in isolated Treg population.
Material and Methods
PBMC Isolation and Culture
PBMC isolation was performed on two EDTA tubes of 8 mL from three healthy donor blood (n=3), using a density gradient with lymphocyte separation medium (Eurobio, Les Ulis, France). PBMCs were washed twice in phosphate buffered saline without calcium and magnesium (PBS -/-) before counting. PBMCs activated or not with anti-CD3 (Miltenyi Biotec, Bergisch Gladbach, Germany) and anti-CD28 (Invitrogen, Carlsbad, CA, USA) at 0.5mg/ml were cultured during 48 hours in RPMI 1640 medium complemented with 1% NEAA, 1% NaPy, HEPES, 0.1% Gentamycin, 0.1% β- Mercaptoethanol (Life technologies, Carlsbad, CA, USA) and 10% human serum AB (SAB) (Sigma Aldrich, Saint-Louis, Missouri, USA)). After 48 hours, PBMCs were harvested, 10 million are put in 1ml of TRIzol reagent (Ambion, Carlsbad, CA, USA) and stored at -80°C until further use.
Treg Isolation
Human blood bags were collected from healthy adult donors at EFS (Etablissement Francais du sang). Treg were isolated from PBMC using Treg isolation kits (Miltenyi Biotec, Bergisch Gladbach, Germany). Obtained purity was over 70%.
RNA Extraction and Reverse Transcription
PBMC total RNA was extracted using TRIzol reagent method according to manufacturer’s instructions. Treg total RNA was extracted using NucleoSpinTM RNA Plus Kit (Macherey Nagel, Hoerdt, France). RNA concentrations were measured using a Nanodrop (ThermoScientific, Waltham, MA, USA). Treg RNA quality was tested using RNA 6000 Nano Kit (Agilent Technologies, Santa Clara, CA, USA) with Bioanalyzer© (Agilent Technologies, Santa Clara, CA, USA). All RNA integrity number were above 8,5. Reverse transcription was performed using SuperScript™ VILO IV™ Master Mix with ezDNase™ Enzyme (ThermoFisher Scientific, Waltham, MA, USA) on 1μg of total RNA. A DNA removal step is performed using ezDNase™ before the reverse transcription.
Selection of Interest Housekeeping Genes
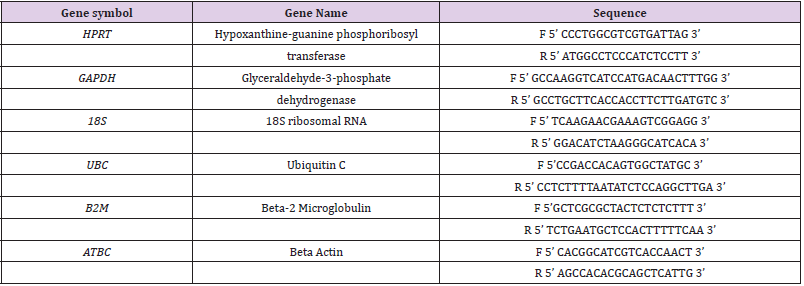
The selection of five housekeeping genes (HPRT, GAPDH, 18S, UBC, B2M) is based on the literature (Table 2). They belong to different functional classes and are reported to be used in genomic studies of immune cells. Primers were designed using FASTA sequences of each gene in Primer Blast (NCBI-NIH) and synthetized by Eurogentech (Liège, Belgium)
Real Time PCR
The RT-PCR reactions were performed, for selected Hk genes (Table 2), according to the manufacturer’s instructions using 2X MESA GREEN qPCR MasterMix Plus for SYBR® 258 Assay (Eurogentech, Liege, Belgium), 96 well qPCR plate (Sarstedt, Hampton, NH, USA), optical seal (Dutcher, Brumath, France) and the Mx3005PTM sequence detection system (Agilent technologies, Santa Clara, CA, USA). In each reaction, 10ng of reverse transcripted RNA (based on initial RNA concentration) was used. All primers were used at 400nM in a 20μL reaction. Quantitative analysis was made based on the cycle threshold (Ct) value for each well and calculated using MxPro software (Agilent Technologies, Santa Clara, CA, USA).
Statistics
The validation of expression stability was statistically tested by R and by BestKeeper methods. BestKeeper algorithm uses raw Ct values and calculates the standard variations (SD and CV). Unstable genes show SD > 1. The most stable HK gene is determined with the correlation coefficient (r) of their expression compared to the BestKeeper Index, which is the geometric mean of Ct values of the highly correlated candidate reference genes. [8,9]. R statistical analysis were done using two Way ANOVA tests.
Data Base-Analysis
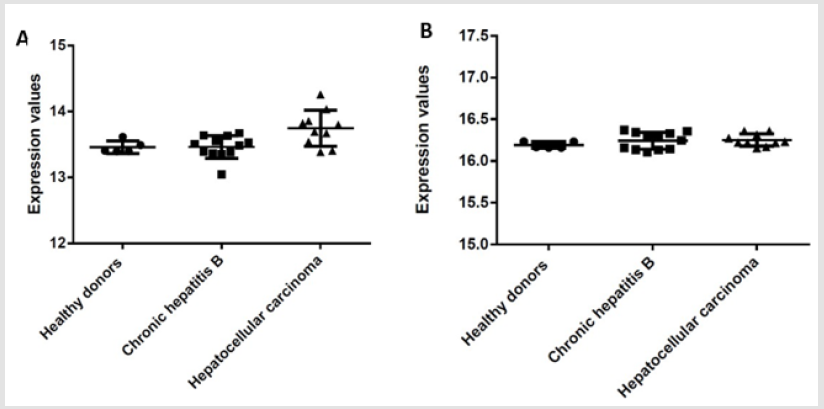
The study shows data obtained in GPL570 data set from Gene Expression Omnibus (GEO). This data include 5 healthy donors, 12 patients with chronic hepatitis B and 10 hepatocellular carcinoma patients.
Results
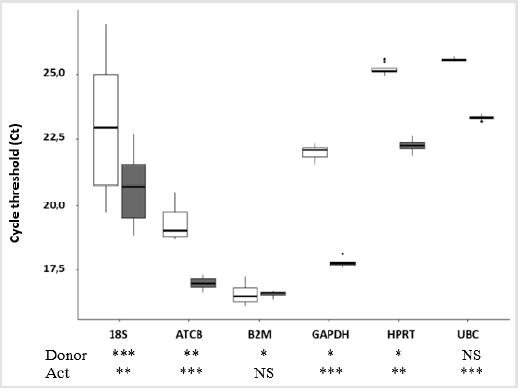
Housekeeping gene expression levels were measured by realtime PCR on 3 healthy donors, and expressions stabilities were tested using both R and BestKeeper algorithms. The ranking of these Hk genes, from the less to the most stable is 18S, ATCB, GAPDH, HPRT, UBC and B2M. Indeed, 18S expression variation between donors is more than 3 Ct, and than 2 Ct regarding the activation state (Figure 1). ATBC is rather stable regarding different donors as variation is less than one Ct, but there is a higher difference, more than 2 Ct, between activated and non-activated PBMCs (Figure 1). GAPDH follows the same trend with nearly no expression variation between donors but with more than 4 Ct of variation following activation (Figure 1). In our results, the rankings of HPRT and UBC is difficult because of their very similar profiles: both are very stable between donors but shows a variation of 2 Ct between activated and non-activated conditions (Figure 1). Nevertheless, statistical analysis by 2-way ANOVA show a weakly significative difference between donors for HPRT compared to the non- significative difference for UBC. Moreover, BestKeeper analysis show power of 1.88 for UBC and of 2.26 for HPRT, as power value must be under 2 for good Hk genes, UBC seems to be better candidate (Table 1). Finally, the most reliable Hk gene in this study is B2M. Indeed, regardless of the activation state the gene expression is very stable, without any significant differences in Ct outcome (Figure 1). Moreover, regarding BestKeeper results, B2M is the only gene with a standard deviation under 1, as recommended (SD = 0.25 CP), and also the gene with the highest p-value [9]. Finally the power is under 2 (Power [x-fold] = 1.07) which is recommended for a good Hk gene as the efficiency of qPCR is set at 2 [9] (Table 1).
Figure 1: Housekeeping gene expression depending on donor or activation state (non-activated in white, activated with anti- CD3/anti-CD28 in grey) analysed by 2 ways ANOVA on R, Ns = no significant difference, * = P ≤ 0.05, ** = P≤ 0.01, *** ≤ 0.001. (n =5).
Table 1: Primers sequences used for the selected housekeeping genes. F is for forward primers and R is for reverse primers.
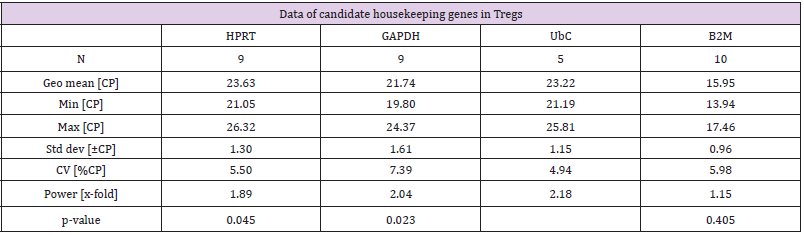
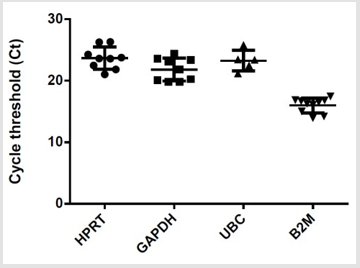
To complete these results a data base analysis has been carried out on 27 patients with different profiles regarding B2M and UBC. Among them, 5 healthy donors, 12 patients with chronic hepatitis B and 10 hepatocellular carcinoma patients [10]. This data-based analysis confirms our results by showing a variation lower than 1Ct for B2M regardless of patients or state of health (Figure 2A). Moreover, it also reinforces UBC results showing nearly no difference between donors but small variations between healthy donors and hepatocellular carcinoma patients (less than 2 Ct) (Figure 2B) To go further, the 4 genes with the least significative difference between donors have been selected to be test on healthy donor primary Treg. In this results B2M expression is stable with a difference under 1 Ct between the 9 donors (Figure 3) with a standard deviation equal to 0.96 (Table 3). Higher variation is observed for HPRT, GAPDH and UBC with respectively standards deviation equal to 1.3; 1.61 and 1.15 (Table 3). Moreover, the power of HPRT and B2M is under 2 as it is recommended for good Hk gene (Table 3).
Figure 3: Housekeeping gene expression in human regulatory T cells. (HPRT, GAPDH n =9, B2M n= 10; UBC n=5).
Discussion
Real-time PCR is a very sensitive and robust quantification method for gene expression analysis. Nonetheless, many factors can impact results, that’s why robusts Hk genes are essential for reliable gene expression quantification . Besides, , as PBMCs includes different cell type, the population holds lots of various information. It is important for us to identify strong Hk gene since PBMC are the more often used sample, easy to collect by small blood collection) for clinical trial follow-up on immune system. Large ATCB expression variation could be explained , since during activation there is a variation of the nucleo-cytoplasmic ratio [5]. The observations regarding GAPDH expression also correlate with other findings, which shows a higher variation in expression, more than 15-folds in 72 different human tissues [11]. Furthermore, the activation mechanism chooses here (anti-CD3, anti-CD28) specifically target T lymphocyte activation by mimicking immunological synapse coactivators. Because of PBMC population heterogeneity, this kind of activation could act indirectly on other cell type activation, notably by cytokine secretion.
Conclusion
Here B2M is the best candidate with stable expression level regardless of the donor and activation state. This result is reinforced by statistical data obtain with Bestkeeper and by databased analysis in PBMCs of cancer patients. Thus, B2M is the only one to complete all the recommendations and is the best candidate for internal control of gene expression in RT-qPCR. This result is coherent since B2M as part of MHC I is equally express on every cell, independently of immune cell activation. Nevertheless, UBC can be also used, as there is no significative difference between donors, confirmed in Treg population. Finally results on HPRT and GAPDH expression in Treg reinforce results on PBMC regarding only few variation depending on donors, that makes these genes usable to compare RT-PCR datas between different donors. Although our results clearly indicate only one perfect candidate, the finding of one other Hk gene is still needed to get best accuracy for PBMC RTPCR and match with the MIQE guideline recommendations
Declarations
Ethics Approval and Consent to Participate
Ethics approval was obtained for this paper. Human blood samples were collected from adult healthy donors after obtaining written consent in accordance with the approval of the Institutional Review Board at the Biology Institute of Lille (DC-2013-1919).
Consent for publication
Not applicable.
Availability of Data and Materials
Not applicable.
Competing Interests
The authors declare that they have not competing interests.
Funding
This project has received financial support from the Société d’Accélération du Transfert de Technologie Nord (SATT Nord), Immune InsighT. The research of CM was supported by the Agence Nationale Recheche Technologie (ANRT).
Author’s Contribution
CM performed all experimentations and participated to the writing of the publication; CI participated to experimental supervision; Dr OM and Pr ND participated to the design of the study and to the writing of the publication. All authors have read and approved the manuscript.
Acknowledgement
Not Applicable.
References
- Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, et al. (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75(2‑3): 291‑29
- Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, et al. (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. BioTechniques Juill 37(1): 112‑11
- Warrington JA, Nair A, Mahadevappa M, Tsyganskaya M (2000) Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiological Genomics 2(3): 143‑14
- Stephen A Bustin, Vladimir Benes, Jeremy A Garson, Jan Hellemans, Jim Huggett, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin chem 55(4): 611-622.
- Hao Zhang, Zi shu Guan, Shi he Guan, Kai Yang, Ying Pan, et al. Identification of Suitable Candidate Reference Genes for Gene Expression Analysis by RT- qPCR in Peripheral Blood Mononuclear Cells of CHB Patients. Clin lab 62(1-2): 227-234.
- Hendrik J M de Jonge, R S Fehrmann, E S Bont, Robert M, F Gerbens, et al. (2007) Evidence Based Selection of Housekeeping Genes. Plos One 2(9): e898.
- Zhang X, Ding L, Sandford AJ (2005) Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol 6: 4.
- Ewa Usarek, Anna Barańczyk Kuźma, Beata Kaźmierczak, Beata Gajewska, Magdalena Kuźma Kozakiewicz (2017) Validation of qPCR reference genes in lymphocytes from patients with amyotrophic lateral sclerosis. Plos One 12(3): e0174317.
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnology Letters 26(6): 509‑5
- Kam M Hui (2014) Gene expression profiling of PBMC from normal individuals, chronic hepatitis B carriers and hepatocellular carcinoma patients.
- Robert D Barber, Dan W Harmer, Robert A Coleman, Brian J Clark (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21(3): 389-395.

 Research Article
Research Article