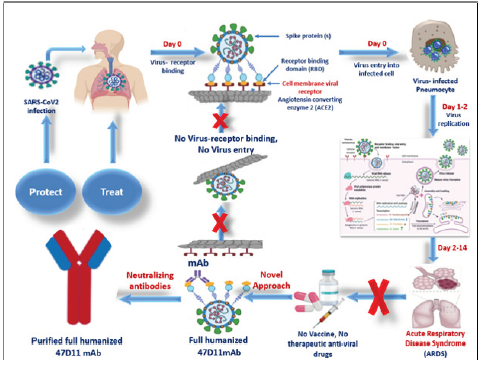
Abstract
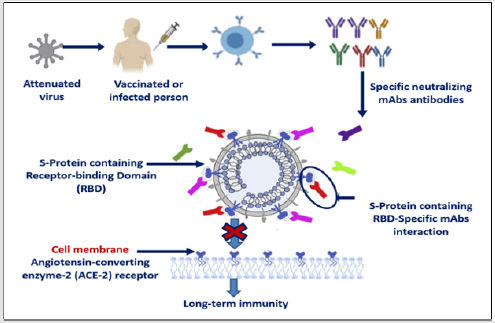
SARS-CoV-2 was appeared firstly in Wuhan, China in December 2019, then spread rapidly to the other countries all over the world, caused a novel Coronavirus-2019 (COVID- 19). The World Health Organization was branded COVID-19 as a global health emergency. After infection, virus entry the host cell through spike proteins (S)- angiotensin converting enzyme 2 (ACE2) cell membrane receptor via their S-Binding domain, then virus replication caused acute respiratory disease syndrome (ARDS). Till now there was no vaccines or anti-viral drugs for coronavirus infection. One effective treatment is the use of human monoclonal antibodies (mAbs) which are engineered to target and block cell surface receptor. mAbs could be given to people in the early stages of COVID-19 as a therapeutic, or used prophylactically to give immediate, long-term immunity to vulnerable people such as healthcare workers. The neutralizing humanized 47D11 monoclonal antibody targets a common epitope on SARS-CoV2 virus and may offer potential for prevention and treatment of COVID-19.89U (Graphical Abstract 1).
Key Point Summaries
a) The initial process for SARA-CoV-2 replication process is binding of S protein to
angiotensin- converting enzyme 2 (ACE2), allowing virus to enter and infect host
cell.
b) Neutralizing antibodies can alter the course of infection in the infected host.
c) Monoclonal antibodies (mAbs) block virus-ACE2 interaction and inhibit virus
entry.
d) mAbs supporting virus clearance or protect an uninfected host that is exposed to
the virus.
e) Purified humanized monoclonal antibody 47D11 was found to be able to block
the infection of SARS-CoV-2.
f) Understanding the mechanisms of neutralizing monoclonal antibodies efficiency,
will provide respected implications in prevention and/or treatment of COVID-19
infection.
Keywords: COVID-19; Immunotherapy; Full-Length Humanized Monoclonal Antibodies; ACE Inhibitor
Abbreviations: ACE2: Angiotensin-Converting Enzyme 2; ARDS: Acute Respiratory Disease Syndrome; CoVs: Coronaviruses; CTL: Cytotoxic T Lymphocyte; E: Envelop Protein; HE: Hemagglutinin; ICTV: International Committee of Taxonomy of Viruses; mAbs: Monoclonal Antibodies; M: Membrane Protein; N: Nucleoprotein; S: Spike Protein; RBD: Receptor Binding Domain; VLP: Virus Like Particles
Introduction
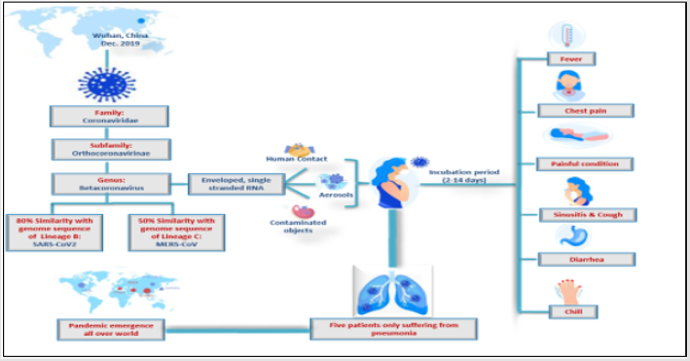
The novel Coronavirus was firstly appeared in Wuhan, China in December 2019 when patients who showed signs of fever, cough and dyspnea with acute respiratory syndrome (ARDS) were reported with unknown viral infection, five patients of all suffering from pneumonia, showed virus genome sequence of previously unknown coronavirus (beta-CoV) strain [1]. The emergence of viral infection spread rapidly caused worldwide epidemic respiratory disease COVID-19 [2]. Coronaviruses (CoVs) are enveloped viruses of the family Coronaviridae (Coronavirinae subfamily), singlestranded RNA genome (26- 32 kb) and positive-sense viruses, that infect host on a wide range, replicate in the cytoplasm of infected cells, and induced diseases ranging from common cold to sever fatal complaints [3]. The novel virus was initially named “COVID- 19” that was changed to “SARS-Cov- 2” by the International Committee of Taxonomy of viruses (ICTV), as it showed 80% similarities to the sequence of SARS- like coronaviruses (SARS-Cov) and 50% to the sequence of MERS- like coronaviruses (MERS-CoV) [4]. Most nations are doing their best efforts for prevention and control strategies, but till now there was no vaccines or anti-viral drugs for coronavirus infection [5]. Understanding of phenotypic and genetic structure of COVID-19 is important for production of vaccines and anti-viral drugs [6] (Figure 1).
Figure 1: Schematic diagram illustrating Coronavirus disease 2019 (COVID-19): Virology and Epidemiology.
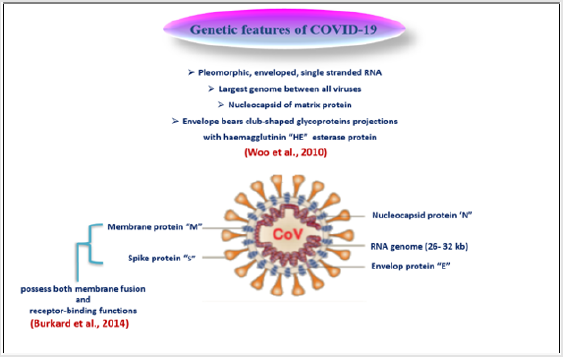
Genetic Features of COVID-19
COVID-9 is pleomorphic enveloped, single stranded RNA within a nucleocapsid composed of matrix protein, and its envelope bears club-shaped glycoprotein projections in addition to hem agglutinin (HE) esterase protein in some coronaviruses [7]. Coronaviruses possess the largest genomes between all RNA viruses. Remarkable features, including a unique N-terminal fragment within spike protein, and genes of these four main structural proteins in all coronaviruses occur in the 5-3 order as large spike protein (S), envelop protein (E), membrane protein (M), and nucleoprotein (N), are encoded by ORFs 10, 11 on the one third of the genome near the 3- terminal [8]. These structural proteins are responsive for viral replication and maintenance of viral genome [9]. It plays a vital role in the intracellular formation of virus particles without requiring S protein [7]. Rendering to genetic features of COVID- 19, it is less like SARS- CoV (about 80%) and MERS- CoV (about 50%), moreover the arrangement of N, E, and M proteins between the three beta coronaviruses are different [2] (Figure 2).
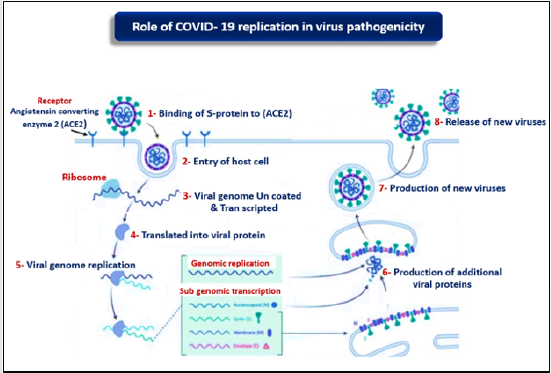
Role of COVID- 19 Replication in Virus Pathogenicity
The initial process for COVID- 19 replication process is binding of S protein to angiotensin- converting enzyme 2 (ACE2), allowing COVID- 19 to enter and infect cell, then the S protein must be primed by protease enzyme called TMPRSS2 to complete cell entrance [10]. After invasion of COVID- 19 the host cell, the genome is uncoated, Tran scripted and Translated, then continuous and unconscious genome synthesis by the viral replicate, and more protein complex encoded by the 20- kb replicase gene [11]. Coronavirus replicase complex is composed of up to 16 viral subunits and a number of cellular proteins, and engagement various RNA processing enzymes that are present in other RNA viruses and include assumed sequence endoribonuclease, 3- to - 5 exoribonuclease, 2 - O- ribose methyltransferase, ADP ribose 1 - phosphatase, and in a subset of group 2 COVs, cyclic phosphodiesterase activities [12]. Cell membrane and RNA assembled proteins are incorporated as mature particle forms by pudding from the internal cell membranes [13] (Figure 3).
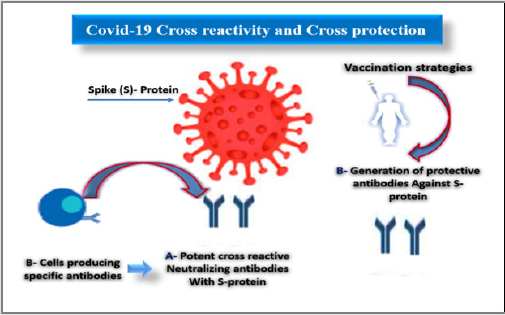
COVID- 19 Cross-Reactivity and Cross-Protection
Cross- reactivity means the reaction between an antibody and an antigen. Cross- Protection is a state in which, infection with a mild virus or viroid strain protects from subsequent infection from a severe strain of the same virus [14]. In recent decades, more efforts are ongoing and have been directed to develop vaccines against human COVID- 19 infection, however cross- protection induced by these vaccines still a restrictive factor as a result of their widespread sequence multiplicity [15]. During the recent threats, immunotherapeutic, and anti-drugs have been explored for their safety, efficacy, and potency [16] (Figure 4).
Vaccination Strategies Against Human COVID-19
Development of COVID- 19 vaccines are based on many strategies; S-protein-based strategies, involved targeting the surface-exposed spike (S) glycoprotein or S protein as the major inducer of neutralizing antibodies, as use of full-length S protein or S1-receptor-binding domain (RBD) and their expression in virus-like particles (VLP), or viral vectors [17]. The major role induced by S protein in induction of protective immunity during infection with SARS-CoV is producing neutralizing-antibodies and T-cell responses [18]. Recently, immuno-informatics was used to recognize significant cytotoxic T lymphocyte (CTL) and B-cell epitopes in SARS-CoV-2 S protein, and the molecular dynamics simulations were used to study the interactions between these epitopes and their corresponding MHC class I molecules, and it was found that the CTL epitopes bind with MHC class I peptidebinding sits through several interactions, therefore indicating their possibility for generating immune responses (Baruah and Bose, 2020).
Therapeutic Monoclonal Antibodies (mAbs) Targeting COVID-19
A novel approach to increase humoral protection against emerging CoVs, using monoclonal antibodies (mAbs) which may play an active role in COVs control. Monoclonal antibodies (mAbs) are produced by B cells and specifically target specific domains in MERS-CoV S protein [19]. Monoclonal antibodies have shown their therapeutic efficacy for several viruses, targeting vulnerable sites on viral surface proteins [20]. Köhler, et al. [21] in 1975 have introduced the hybridoma technique to acquire pure mAbs in great amounts, stimulating the basic research and possibility for clinical use [22]. Neutralizing antibodies mainly target the trimeric spike (S) glycoproteins- containing RBD on the viral surface that mediate entry into host cells. Effective neutralizing antibodies frequently target the receptor interaction site in S1 [19]. The spike proteins of SARS-CoV-2 and SARS-CoV are structurally very similar with 77.5% similarity in amino acid sequence [23] and regularly bind the human angiotensin converting enzyme 2 (ACE2) protein as a host receptor via their S-Binding domain [24]. Therapeutic antibodies typically have long serum half-lives, which allow for infrequent administration, every other week or less frequently [25] (Figure 5).
Generation of H2L2 mAbs Against COVID-19
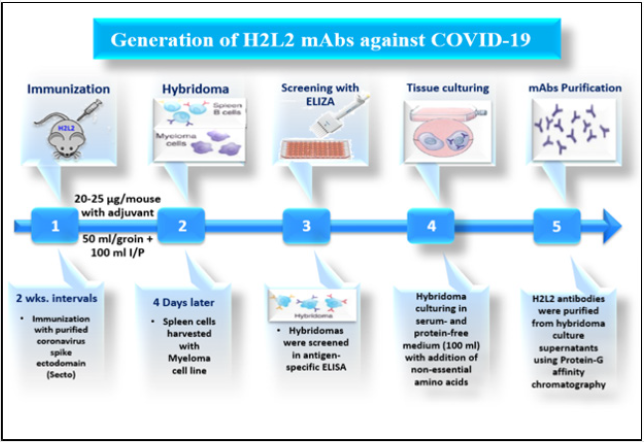
H2L2 mice were sequentially immunized in 2 weeks intervals with purified Coronavirus spike ectodomains (Secto) of different CoVs in the following order: HCoV-OC43, SARS-CoV, MERS-CoV, HCoV-OC43, SARS-CoV, and MERS-CoV. Antigens were injected at 20-25 μg/mouse using freshly prepared adjuvant. Injections were done subcutaneously into the left and right groin each (50 μl) and 100 μl intraperitoneally. Four days after the last injection, spleen and lymph nodes are harvested, and hybridomas made by standard method using SP 2/0 myeloma cell line. Hybridomas were screened in antigen-specific ELISA and those selected for further development, subcloned a in serum- and protein-free medium (100 ml) with addition of non-essential amino acids. H2L2 antibodies were purified from hybridoma culture supernatants using Protein-G affinity chromatography and stored at 4˚C until use [26] (Figure 6).
Figure 6: Dynamic diagram showing Generation of therapeutic H2L2 mAbs against Covid-19 using mouse hybridoma technique.
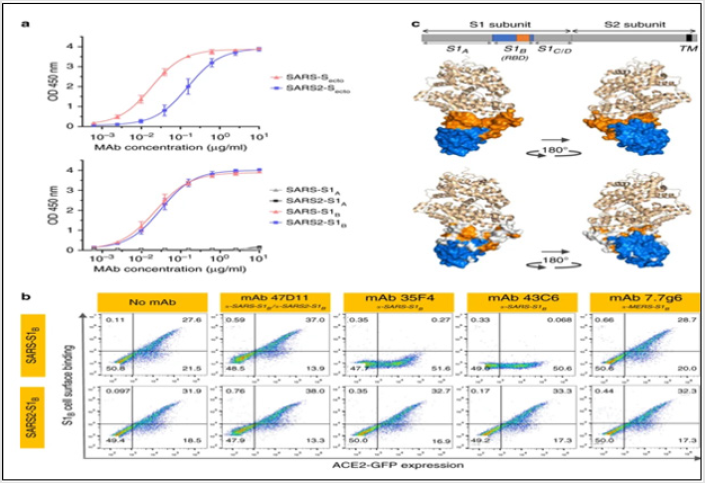
The neutralizing 47D11 mAb binds SARS1-S and SARS2-S RBD
In order to identify SARS-CoV-2-neutralizing antibodies, ELISA- (cross) reactivity was assessed of antibody-containing supernatants of a collection of 51 SARS-S hybridoma’s derived from immunized transgenic H2L2 mice that encode chimeric immunoglobulins with human variable heavy and light chains and constant regions of rat origin. Four of 51 SARS-S hybridoma supernatants displayed ELISAcross- reactivity with the SARS-S1 and SARS2-S1 subunits. 47D11 exhibited cross-neutralizing activity of SARS-S and SARS2-S pseudo typed VSV infection. The chimeric 47D11 H2L2 antibody was reformatted to a fully human immunoglobulin, by cloning of the human variable heavy and light chain regions into a human IgG1 isotype backbone [27] (Figure 7). Prior to cell binding, S1B was mixed with mAb (mAbs 47D11, 35F4, 43C6, 7.7G6, in H2L2 format) with indicated specificity in a mAb:S1B molar ratio of 8:1. Cells are analyzed for (ACE2-)GFP expression (x axis) and S1B binding (y axis). Percentages of cells that scored negative, single positive, or double positive are shown in each quadrant [27].
Figure 7: The neutralizing 47D11 mAb binds SARS-S and SARS2-S RBD.
a. ELISA-binding curves of 47D11 to Secto (upper panel) or S1A and S1B (RBD: receptor-binding domain) (lower panel) of
SARS-S and SARS2-S coated at equimolar concentrations. The average ± SD from two independent experiments with technical
duplicates is shown.
b. Interference of antibodies with binding of the S-S1B of SARS-CoV and SARS-CoV-2 to cell surface ACE2-GFP analyzed
by flow cytometry.
Production of Full Humanized 47D11mAbs Using pTRIOZ Plasmidsc
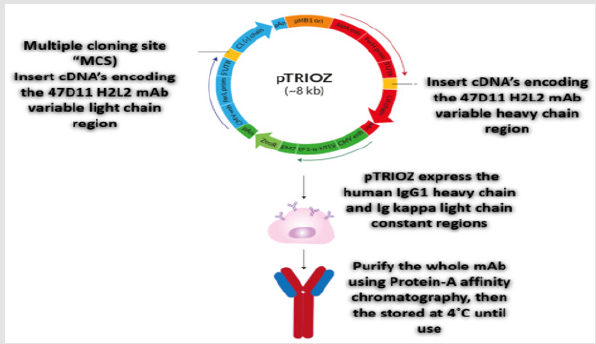
pTRIOZ plasmid used specifically for successful production of whole recombinant monoclonal antibodies (mAbs). Immunized H2L2 mice have been engineered to produce a fully human antibody (instead of mouse). For humanized monoclonal antibody 47D11 production, the cDNA’s encoding the 47D11 H2L2 mAb variable regions of the heavy and light chains were cloned into expression plasmids containing the human IgG1 heavy chain and Ig kappa light chain constant regions respectively. Both plasmids contain the interleukin-2 signal sequence to enable efficient secretion of recombinant antibodies. Protein-A affinity chromatography was used for purification of Human antibodies, then the purified antibodies were stored at 4˚C until use [28-31] (Figure 8).
Figure 8: Schematic review of the pTRIOZ collection designed specifically for high-yield production of whole recombinant monoclonal antibodies (mAbs).
References
- Huang C, Wang Y, Li X, Lili R, Jianping Z, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395(10223): 497-509.
- Lu H (2020) Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 14(1): 69-71.
- Gorbalenya AE, Baker SC, Baric RS, Groot PR, Drosten C, et al. (2020) Sever acute respiratory syndrome-related coronavirus: the species and its viruses- a statement of the coronavirus study group. Nature Microbiology 5: 536-544.
- Kuldeep D, Khan S, Ruchi T, Maryam D, Yashpal SM, et al. (2020) COVID-19, an emerging corovirus infection: advances and prospects in designing and developing vaccines, immunotheapeutics, and therapeutics. Human Vaccins&Immunother 16(6):1232-1238.
- Pillaiyer T, Meennakshisundaram S, Manickam M (2020) Recent discovery and development of inhibitors targeting coronavirus. Drug discov Today 25(4): 668-688.
- Leila M, Sorayya G (2020) Genotype and Phenotype of Covid-19: Their roles in pathogenesis. J Microbiol Immunol Infect.
- De Haan CAM, Kuo L, Masters PS, Vennema H, Rottier PJM (1998) Coronavirus particle assembly: Primary structure requirements of the membrane protein. J Virol 72(8): 6838-6850.
- Woo PCY, Huang Y, Lau SKP, Yuen KY (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2(8): 1804-1820.
- Van Boheemen S, De Graaf M, Laubr G, Bestebroer TM, Raj VS, et al. (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6): e00473-512.
- Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, et al. (2020) The origin, transmission and clinical therapies on coronavirus disease 2010 (COVID- 19) outbreak- an update on the status. Mild Med Res 7(1): 11.
- Ziebuhr J (2005) The Coronavirus Replicase. Coronavirus Replication and Reverse Genetics. Current Topics in Microbiology and Immunology 287: 55-94.
- Almazan F, DeDiego ML, Galan C, Escors D, Alvarez E, et al. (2006) Construction of a svere acute respiratory syndrome coronavirus infection Cdna clone and a replicon to study coronavirus RNA synthesis. J Virol 80 (21): 10900-10906.
- Mcintosh K, Peiris JSM (2009) Coronavirus. Clinical virology (3rd), American Society of Microbiology, pp. 1155-1171.
- Ziebell H, Carr JP (2010) Cross-protection: a century of mystery. Advances in virus research 76: 211-264.
- Graham RL, Donaldson EF, Baric RS (2013) A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11(12): 836-848.
- Cyranoski D (2020) This scientist hopes to test coronavirus drugs on animals in locked-down Wuhan. Nature 577(7792): 607.
- Jiang S, He Y, Liu S (2005) SARS vaccine development. Emerg Infect Dis 11(7): 1016-1020.
- Du L, He Y, Zhou Y, Liu S, Zheng BJ, et al. (2009) The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3): 226-236.
- Widjaja I, Wang C, Haperen RV, Gutiérrez Álvarez J, Van Dieren B, et al. (2019) Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerging Microbes Infect 8(1): 516-530.
- Rockx B, Corti D, Donaldson E, Sheahan T, Stadler K, et al. (2008) Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J. Virol 82(7): 3220-3235.
- Kohler G, Milstein C (1975) Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256(5517): 495-497.
- Goo J, Jeong Y, Park YS, Yang E, Jung DI, et al. (2020) Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein. Virus Res 278: 197863.
- Gui M, Song W, Zhou H, Xu J, Chen S, et al. (2017) Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27(1): 119-129.
- Li W, Michael JM, Natalya V, Jianhua S, Swee KW, et al. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426(6965): 450-454.
- Souders CA, Nelson SC, Wang Y, Crowley AR, Klempner MS, et al. (2015) A novel in vitro assay to predict neonatal Fc receptor-mediated human IgG half-life MAbs 7(5): 912-921.
- Lu R, Zhao X, Li J, Yang B, Wu H, et al. (2020) Genomic characterization and epidemiology of 2019 novel coronavirus. Implications for virus origins and receptor binding. Lancet 395(10224): 566-574.
- Meulen J, Van den Brink EN, Poon LLM, Marissen WE, Leung CSW, et al. (2006) Human Monoclonal Antibody Combination against SARS Coronavirus: Synergy and Coverage of Escape Mutants. PLoS Med 3(7): e237.
- Wang C, Li W, Dubravka D, Nisreen MA, Haperen ORV, et al. (2020) A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 11(1): 2251.
- Christine B, Monique HV, Oliver W, Sander IK, Frank J, et al. (2014) Coronavirus cell entry occurs through the endo/lysosomal pathway in a proteolysis-dependent manner. PlosPathog 10(11): e1004502.
- Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky A (2020) Covid-19 spike- host cell receptor GRP78 binding site prediction. J Infect 80(5): 554-562.
- Spiga O, Bernini A, Clutti A, Chiellini S, Menclassi N, et al. (2003) Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys, Res. Commun 310(1): 78-83.

 Review Article
Review Article