Abstract
Endometriosis is a chronic gynecological inflammatory disease characterized by the presence of functional endometrial glands and stroma outside of the uterine cavity. It is an estrogen-dependent disease affecting 7-10% of women of reproductive age. Laparoscopic diagnosis remains the gold standard approach, providing that the risks these minimally invasive surgical procedures are understood and accepted by the patients. The aim of this study was to compare urine proteomic profiles in patients with endometriosis versus controls, trying to identify a panel of specific proteins representing novel non-invasive biomarkers of endometriosis. Eight 30-40 years old women with laparoscopically verified ovarian endometriotic cysts were enrolled. Five fertile women with non-endometriotic ovarian cystsand no visual signs of peritoneal endometriosis served as control group. Morning urine were collected in the day of the scheduled surgery and processed to be analyzed by proteomic. Eleven proteins were increased in endometriosis, while prothrombin was reduced. Seven proteins were found to fall into one large protein-protein interaction network, mainly involved with inflammation. This is the first report on the application of quantitative proteomics in the search of an array of urinary proteins, which may guide further studies for the validation of novel non-invasive biomarkers of endometriosis.
Keywords: Endometriosis; Urine; Markers; Non-Invasive; Proteins; Proteomics
Introduction
Endometriosis is a chronic gynecological inflammatory disease characterized by the presence of functional endometrial glands and stroma outside of the uterine cavity. Endometriosis affects 7-10% of women of reproductive age, 60% of women with pelvic pain and up to 50% of women with infertility [1]. The spectrum of symptoms includes dysmenorrhea, dyspareunia, chronic pelvic pain, dyschezia, dysuria. However, recognition and conclusive diagnosis is only made on the average approximately 9 years following the beginning of the disease [2]. Early detection of the disease would allow patients earlier options for treatment, i.e. a sooner medical or surgical treatment and follow-up. The current gold standard for the diagnosis of the disease requests laparoscopic evaluation and biopsy of the visualized lesions [3]. Although laparoscopy is a minimally invasive procedure, it requires general anesthesia, developed surgical skills and it has a high procedural cost. In addition, laparoscopy is associated with a, although rare, risk of potential intra-operative or post-operative complications [4,5]. The development of high-throughput technologies, such as proteomics, has led to an early diagnosis of medical diseases through the identification of proteins linked to different pathological conditions [6]. In previous studies we have demonstrated that cervical mucus is a source of putative protein biomarkers [7]. We have also characterized a pattern of cervical mucus proteins involved in the pathophysiology of endometriosis [8], which might represent putative non-invasive markers of endometriosis. Urine proteomic analysis is a valuable source to identify specific proteins and peptides. Compared to cervical mucus collection, urine proteomic analysis has the advantage to obtain proteins in a non-invasive, simple method and in a large quantity.
It has been defined as a fluid biopsy of the urogenital tract, which provides considerable information about these organs [9]. Consequently, many changes in urogenital tract function may be detected by urinary proteomic analysis. Currently, urinary proteomics is mainly used as diagnostic tool to identify biomarkers of kidney diseases, such as IgA nephropathy, renal cell carcinoma, and to detect biomarkers for chronic allograft dysfunction after kidney transplantation [10]. In addition, urinary proteomics have been used to study a variety of diseases, such as obstructive sleep apnea and obesity [11] and brain [12] and cardiovascular diseases [13]. Protein metabolites excreted in urine may reflect the state of the body and serve as informative biomarkers for some diseases. Through urinary proteomics, the molecule metabolites in urine can be comprehensively analyzed (metabolomics). Urinary metabolomics has been widely used in the study of various diseases, including diabetic nephropathy [14], chronic heart failure [15] and offers potential early diagnosis of ovarian and breast cancer [16]. Previous studies have been published attempted to identify the proteomic profiling of endometriosis in urine samples. Using two-dimension gel separation coupled with mass spectrometry, Tokushige et al. [17] demonstrated the unique presence of a highly up-regulated protein, the CK-19, in the urine of women with endometriosis compared to a control group of women without endometriosis. Recently, urine peptide biomarkers have been searched using magnetic beads-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDITOF- MS). Wang et al. [18] identified in urine samples of patients with endometriosis a specific cluster of peptides. However, an exhaustive identification of the list of putative urine markers of endometriosis has not been still provided. The aim of this study was to identify, in urine of patients affected by endometriosis, a panel of candidate proteins, which may be further validated as novel putative biomarkers of this pathological condition.
Materials and Methods
Ethical Approval
The Ethical board of “Fondazione Policlinico “A. Gemelli” approved the protocol (registration number 0019148/16) on May, 5th 2016.
Patients
Eight 30 to 40 years old infertile women, who underwent laparoscopy for ovarian endometriotic cysts, confirmed at histological exam, were enrolled. They were compared with 5 fertile women, with non-endometriotic ovarian cysts, confirmed at histological exam. Patients and controls were all operated upon by a surgeon with a long previous experience in the laparoscopic diagnostic and treatment of endometriosis (R.M.) [1,19,20]. The inclusion criterion for the study was: patients 30 to 40 years old for both groups, with endometriotic or non-endometriotic ovarian cysts. Patients with non-endometriotic ovarian cysts had obtained previous pregnancies. Final diagnosis was obtained by video laparoscopy and was confirmed by histological exam of the cyst (endometriotic or non-endometriotic). The exclusion criteria for both groups were the following: patients with other gynecologic, kidney and systemic diseases (known cancer, diabetes, thyroid disease, cardiovascular disease).
Urine Collection
Urine culture was performed in the week preceding admission
for surgery, to exclude urinary tract infection.
Morning midstream urine samples were collected by patients
and controls after hospitalization in the morning of surgery.
Immediately after urine collection, a tablet of cocktail of protease
inhibitor (Roche-Basel, Switzerland) was added to 10 ml of samples.
Sample Preparation
Ten milliliters of each urine samples were centrifuged at 1.500×g, 10 minutes at 4°C. The supernatant has been collected and stored at -80°C. One milliliter of each urine sample has been added with acetonitrile (4 volumes), vortex mixed and incubated at -20°C overnight. After centrifugation at 10.000×g, 20 minutes at 4°C, the supernatant was discharged and the precipitated pellet were resuspended in 100μL of Urea 6M/Tris 100 mM pH 7.8 buffer. Total protein content was quantified by Bradford assay. 10μg of each sample have been subjected to in-solution digestion protocol as described previously [21]. Briefly, each sample corresponding to 10μg of total protein has been mixed with 100mM of ammonium bicarbonate at pH 8.0 and reduced with 200mM dithiothreitol (DTT, 10mM final; Sigma) for 5 minutes at 100°C, 15 minutes at 50°C, and alkylated with 200mM of iodoacetamide (55mM final; Sigma) in the dark at room temperature for 60 minutes. The samples were left to digest overnight at 37°C by adding 100mM of ammonium bicarbonate (pH 8) with sequencing grade-modified porcine trypsin (1:50, trypsin: protein concentration; Promega). To stop the digestion, the samples have been acidified with aqueous trifluoroacetic acid (TFA/H2O 0.2%vol/vol), immediately frozen, and lyophilized.
LC-MS/MS analysis
For proteomic analysis, the samples have been resuspended in 40μL of aqueous formic acid solution (0.1% v/v) and equal protein quantity (3.5μg) of each sample has been analyzed by Ultimate RSLC nano apparatus coupled to Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Separation experiments have been performed using a Zorbax C18 column (3.5μm particle diameter, 1mm i.d. x 15cm) (Agilent Technologies, Santa Clara, CA) using following eluents: (A) 0.1% (v/v) aqueous formic acid and (B) acetonitrile: water (80:20) with 0.1% aqueous formic acid. The gradient applied, was linear from 0 to 70% of solvent B in 40 min, at a flow rate of 50μl/min. The LTQ Orbitrap Elite has been operated in data dependent mode in which each full MS scan was followed by MS/MS scans where the five most intense multi-charged ions have been dynamically selected and fragmented by collision-induced dissociation (CID) at normalized collision energy of 35%.
Protein Identification and Quantification
Tandem mass spectra have been elaborated by Proteome Discoverer 1.4 software (version 1.4.1.14 licensed by Thermo Fisher Scientific), based on SEQUEST HT cluster as search engine, against Homo Sapiens proteome (UniProtKB/SwissProt release 2018-03). Data have been elaborated with following parameters: trypsin enzyme with maximum of two missed cleavages, fixed carbamidomethylation of cysteine and the oxidation of methionine as variable modification. In order to obtain a reliable identification of the proteins/peptides the following stringent criteria have been used: two unique peptide per protein, high peptide confidence (False discovery rate <1%). A 1% FDR and a minimum of two peptide matches per protein have been the criteria used for protein identification. The dissociated or ‘ungrouping’ of proteins from their respective families has been used during the quantification process to avoid the possible ambiguity associated with different isoforms of the same protein. The label-free quantitative analysis has been performed via Precursor Ions Area Detector Node during the bioinformatic analysis using Proteome Discoverer software. This quantification method has been used to define the relative quantities of all peptides in a sample. The Proteome Discoverer application calculates peptide areas during processing, using them to automatically calculate protein areas for the proteins in the report. It calculates the area of any given protein as the average of the three most abundant distinct peptides identified in the protein.
Bioinformatics and Statistical Analysis
The relative protein level ratios between the group of controls (n=5) and the group of patients (n=8) have been determined from the respective averages of protein abundances expressed for each protein in the two groups. All the proteins detected with a ratio>1.5 (less abundant proteins in patients) or <0.67 (more abundant proteins in patients) have been considered for this study. Proteinprotein interaction network analysis has been performed by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). STRING is a database for predicted protein-protein interactions at EMBL clusters the extracted results from many protein-protein interactions databases, like Mint, BioGrid, etc. It also uses the information from KEGG pathways and Reactome to provide the best annotations for the interactions of one protein [22]. Analysis has been performed considering a high confidence of interactions (0.7), evaluated for experiments, databases, co-occurrence and coexpression. MCL clustering has been performed defining 3 as MCL inflation parameter.
Results
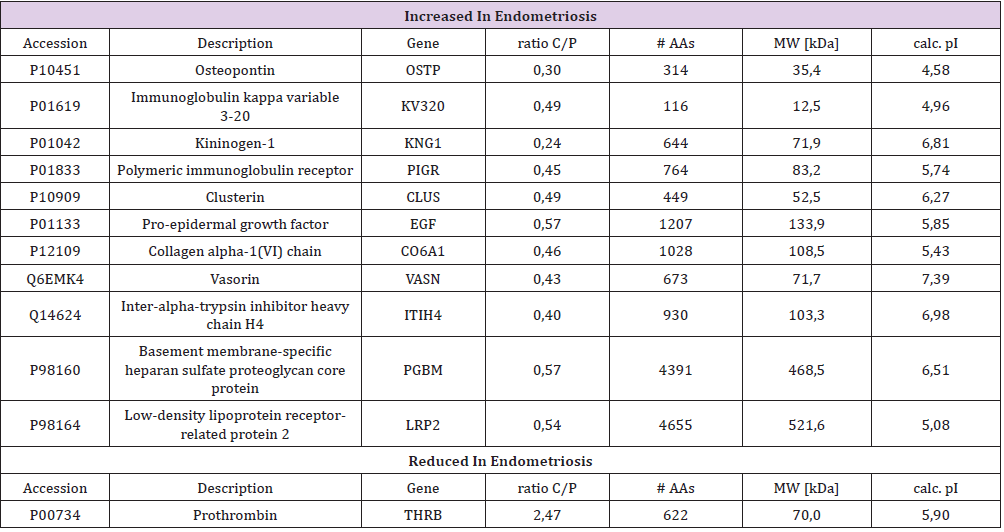
Protein identification led to the characterization in the endometriosis group of 65-92 different urinary proteins per sample in the group of patients with endometriosis and of and 32-77 proteins per sample in the control group. Eleven proteins were found to be increased in patients with endometriosis (Table 1), while one protein (prothrombin) was found to be decreased in patients with endometriosis versus the control group.
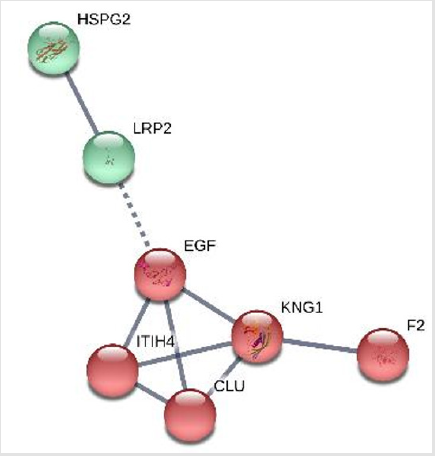
Seven out of the 12 differentially expressed proteins fall into a large protein-protein interaction network based on the prediction results of STRING system (Figure 1). Two clusters have been built, the major one composed by 5 proteins (kininogen 1, Epidermal growth factor (EGF), Inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4), clusterin (CLU) and thrombin (F2), mainly involved with inflammation.
Discussion
In the present study, the human urinary proteomic pattern was investigated for the first time to search for altered molecules in patients with endometriosis. We identified a panel of 12 differentially expressed proteins, which might represent novel putative markers for non-invasive diagnosis of endometriosis. Further high-scale specific studies are needed to confirm these data. Among these 12 proteins, we reported a network of 7 proteins strictly related with inflammation. These proteins represent a signature of the systemic chronic inflammatory status induced by endometriosis. Inflammatory processes have in fact a crucial role in the pathophysiology of endometriosis [23], as suggested by the abnormal levels of immune system cells within the female reproductive tract reported in patients with endometriosis. The immune system cells were moreover found to be dysfunctional in endometriosis [24]. As a consequence after the implantation of endometriotic cells within the abdominal cavity, there is an endometriosis-induced secretion of cytokines, chemokines, nitric oxide, immunoglobulins. Supporting this physio-pathological hypothesis we previously demonstrated that IL-10, although at low doses, exerts its action on endometrial cells in modulating antiinflammatory and anti-apoptotic effects in vitro [25]. In the present study we identified a cluster of proteins linked with inflammatory reactions, constituted by 5 proteins (KNG1, ITIH4, CLU, EGF and F2).
Kininogen 1 is a protein that in humans is encoded by the KNG1 gene. The KNG1 gene uses alternative splicing to generate two different proteins: high-molecular-weight kininogen (HMWK) and low-molecular-weight kininogen (LMWK). HMWK in turn is cleared by the enzyme kallikrein to produce bradykinin. Kallikrein in the presence of HMWK stimulates mononuclear chemotaxis [26] and induces the release of neutrophil elastase. Bradykinin stimulates inflammation, releases prostaglandins and nitric oxide, and enhances microvascular flow and permeability [27]. KNG 1 plays moreover an important role in regulating, at gene level, urokinase plasminogen activator receptor, which is involved in cell migration and proliferation. The increased urinary excretion in endometriosis of KNG1 might so reflect an increase of KNG1 production and release, as part of the inflammatory reaction which is observed in endometriosis. Inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4) is an acute-phase inflammatory response protein that belongs to a super family of protease inhibitors [28]. Winden et al. [29] reported ITIH4 detection in blood up to 3 years prior to the diagnosis of some diseases, such as breast cancer and colorectal cancer, which supports its potential use as a diagnostic biomarker. Since both KNG1 and ITIH4 have been reported in urines as reduced in ovarian cancer at early stage [30], while we reported their increase in ovarian endometriosis, their evaluation might represent a non-invasive marker to be translated in clinical practice for a further evaluation of ovarian disease.
Clusterin (CLU) was previously reported as increased in peritoneal fluid and in cervical mucus in women with endometriosis [31] Clusterin is a ubiquitous protein that is arousing increasing interest owing to its widespread diffusion and multifunctional role. It is composed of two 40 kD subunits (NA1, NA2) encoded by a single gene, which are hold together by disulphide bonds. It is a component of the high-density lipoprotein (HDL) complex, with putative functions in the transport of lipids, apoptosis and protection of cells from stress and, most important, in the regulation of complement activity. There is strong evidence that clusterin plays a role in renal diseases. In fact, clusterin co-localizes with the C5b-9 complex within immune deposits in human membranous glomerulonephritis [32] where it is supposed to act as a regulator of the C5b-9 cell injury. Depletion of clusterin enhances immune glomerular injury in the isolated perfused kidney [33] and clusterin -/- knockout mice are more prone to injury due to immunocomplexes [34]. In 1995 Megalin was identified as the target of the C5b-9 injury in experimental membranous glomerulonephritis [35]. Megalin is the cell receptor for clusterin in many tissues [36], so that the presence of clusterin in glomeruli represents a host limitation to complement injury, as it prevents C5b-9 insertion into cell membrane, where the complement complex competes with clusterin for the access to the same receptor, which is Megalin. Several previous studies reported that complement pathway is frequently altered in endometriosis, with an increase in the levels of C5b-9 component [37]. As a consequence we may speculate that the increase in the levels of clusterin in the urines of patients with endometriosis might reflect the high levels of clusterin, as acute phase protein, present in endometriosis. On the other hand, the higher excretion of both clusterin and megalin in urines of patients with endometriosis might reflect a protective mechanism for kidneys to face the possible injury due to increased level in complement complexes in endometriosis. Further studies are needed to clarify the role of clusterin-megalin system in patients with endometriosis, particularly at the kidney level.
Urinary EGF (epidermal growth factor) has been proposed as a marker for different non-urological diseases [38]. Intriguingly, the EGF system is strictly involved in the pathogenesis of endometriosis [39]. It is known that the expression of EGF system in eutopic endometrium from women with endometriosis varies from the endometrium of healthy women, with significant quantitative and qualitative differences [40]. In addition, a dysregulation of EGF system has been demonstrated in the setting of severe versus mild endometriosis, suggesting functional and biochemical dissimilarities between these two types of endometriosis [41]. As a consequence, the higher urinary EGF levels observed in patients with endometriosis might reflect the dysfunctional EGF system in this disease. Finally, in the present study we reported for the first time the lower urinary prothrombin levels in patients with endometriosis. It might reflect the changes in coagulation status that has been described in patients with endometriosis [42]. Recent evidences in fact have reported higher prevalence of trombophilic disorders in endometriosis patients [43]. The lower levels of prothrombin in urines of endometriosis patients might reflect this specific coagulation status. This is the first reported application of quantitative high-resolution mass spectrometry – based proteomics that detected a specific array of proteins inthe urines of patients with endometriosis. The findings of the present study confirm that endometriosis could be viewed as a disease with systemic inflammatory involvement, as reflected by metabolites excreted in urines. These data call for further confirming studies for the validation of these non-invasive biomarkers of endometriosis.
Acknowledgment
The authors thank Claudia Menaldino (New York, USA) for her kind and careful English editing.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Information
No specific funding was obtained for this project.
References
- Marana R, Lecca A, Biscione A, Muzii L (2012) Endometriosis: The gynecologist’s opinion. Urol J 79(3): 160-166.
- Luisi S, Lazzeri L, Ciani V, Petraglia F (2009) Endometriosis in Italy: From cost estimates to new medical treatment. Gynecol Endocrinol 25(11): 734-740.
- Kennedy S, Bergqvist A, Chapron C, D Hooghe T, Dunselman G, et al. (2005) ESHRE guideline for the diagnosis and treatment of endometriosis. Hum Reprod 20(10): 2698-2704.
- Karaman Y, Uslu H (2015) Complications and Their Management in Endometriosis Surgery. Women’s Heal 11(5): 685-692.
- Chapron C, Vercellini P, Baraka H, Vieira M, Dubuisson J-B (2002) Management of ovarian endometriomas. Hum Reprod Update 8(6): 591-597.
- Duarte TT, Spencer CT (2016) Personalized Proteomics: The Future of Precision Medicine. Proteomes 4(4): 29.
- Grande G, Milardi D, Vincenzoni F, Pompa G, Biscione A, et al. (2015) Proteomic characterization of the qualitative and quantitative differences in cervical mucus composition during the menstrual cycle. Mol BioSyst 11(6): 1717-1725.
- Grande G, Vincenzoni F, Milardi D, Pompa G, Ricciardi D, et al. (2017) Cervical mucus proteome in endometriosis. Clin Proteomics 14(1): 7.
- Bellei E, Monari E, Bergamini S, Pini LA, Aldo T, et al. (2018) Urinary Proteomics in Biomarker Discovery of Kidney-Related Disorders: Diabetic Nephropathy and Drug-Induced Nephrotoxicity in Chronic Headache. EJIFCC 29(4): 290-297.
- Quintana LF, Solé-Gonzalez A, Kalko SG, Bañon-Maneus E, Solé M, et al. (2009) Urine proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol 20(2): 428-435.
- Seetho IW, Siwy J, Albalat A, Mullen W, Mischak H, et al. (2014) Urinary proteomics in obstructive sleep apnoea and obesity. Eur J Clin Invest 44(11): 1104-1115.
- An M, Gao Y (2015) Urinary Biomarkers of Brain Diseases. Genomics Proteomics Bioinformatics. 13(6): 345-354.
- Delles C, Diez J, Dominiczak AF (2011) Urinary proteomics in cardiovascular disease: Achievements, limits and hopes. Proteomics Clin Appl 5(5-6): 222-232.
- Ng DPK, Salim A, Liu Y, Zou L, Xu FG, et al. (2012) A metabolomic study of low estimated GFR in non-proteinuric type 2 diabetes mellitus. Diabetologia 55(2): 499-508.
- Kang SM, Park JC, Shin MJ, Lee H, Oh J, et al. (2011) 1H nuclear magnetic resonance based metabolic urinary profiling of patients with ischemic heart failure. Clin Biochem 44(4): 293-299.
- Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, et al. (2010) Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res 16(23): 5835-5841.
- Tokushig N, Markham R, Crossett B, Ahn SB, Nelaturi VL, et al. (2011) Discovery of a novel biomarker in the urine in women with endometriosis. Fertil Steril 95(1): 46-49.
- Wang L, Liu HY, Shi HH, Lang JH, Sun W (2014) Urine peptide patterns for non-invasive diagnosis of endometriosis: a preliminary prospective study. Eur J Obstet Gynecol Reprod Biol 177: 23-28.
- Marana R, Paparella R, Ferrari S, Pompa G, Paparella P, et al. (2008) Laparoscopic diagnosis of peritoneal endometriosis. Minerva Ginecol 60(6): 552-554.
- Muzii L, Marana R, Angioli R, Bianchi A, Cucinella G, et al. (2011) Histologic analysis of specimens from laparoscopic endometrioma excision performed by different surgeons: does the surgeon matter? Fertil Steril 95(6): 2116-2119.
- Milardi D, Grande G, Vincenzoni F, Giampietro A, Messana I, et al. (2014) Novel Biomarkers of Androgen Deficiency From Seminal Plasma Profiling Using High-Resolution Mass Spectrometry. J Clin Endocrinol Metab 99(8): 2813-2820.
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, et al. (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39(Database issue): D561-568.
- Jiang L, Yan Y, Liu Z, Wang Y (2016) Inflammation and endometriosis. Front Biosci Landmark Ed 21: 941-948.
- Thiruchelvam U, Wingfield M, O’Farrelly C (2015) Natural Killer Cells: Key Players in Endometriosis. Am J Reprod Immunol 74(4): 291-301.
- Mancini F, Milard D, Carfagna P,Grande G, Miranda V, et al. (2018) Low-dose SKA Progesterone and Interleukin-10 modulate the inflammatory pathway in endometriotic cell lines. Int Immunopharmacol 55: 223-230.
- Gallin JI, Kaplan AP (1974) Mononuclear cell chemotactic activity of kallikrein and plasminogen activator and its inhibition by C1 inhibitor and alpha 2-macroglobulin. J Immunol 113(6):1928-1934.
- Kaplan AP, Joseph K, Silverberg M (2002) Pathways for bradykinin formation and inflammatory disease. J Allergy Clin Immunol 109(2): 195-209.
- Zhuo L, Hascall V, Kimata K (2004) Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem 279(37): 38079-38082.
- Opstal-van Winden AWJ, Krop EJM, Kåredal MH, Gast M-CW, Lindh CH, et al. (2011) Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer 11: 381.
- Abdullah-Soheimi SS, Lim BK, Hashim OH, Shuib AS (2010) Patients with ovarian carcinoma excrete different altered levels of urine CD59, kininogen-1 and fragments of inter-alpha-trypsin inhibitor heavy chain H4 and albumin. Proteome Sci 8: 58.
- Konrad L, Hackethal A, Oehmke F, Berkes E, Engel J, et al. (2016) Analysis of Clusterin and Clusterin Receptors in the Endometrium and Clusterin Levels in Cervical Mucus of Endometriosis. Reprod Sci 23: 1371-1380.
- Murphy BF, Kirszbaum L, Walker ID, d’Apice AJ (1988) SP-40,40, a newly identified normal human serum protein found in the SC5b-9 complex of complement and in the immune deposits in glomerulonephritis. J Clin Invest 81(6): 1858-864.
- Saunders JR, Aminian A, Mc Rae JL, O’Farrell KA, Adam WR, et al. (1994) Clusterin depletion enhances immune glomerular injury in the isolated perfused kidney. Kidney Int 45(3): 817-827.
- Rosenberg ME, Girton R, Finkel D, Chmielewski D, Barrie A, et al. (2002) Apolipoprotein J/clusterin prevents a progressive glomerulopathy of aging. Mol Cell Biol 22(6):1893-1902.
- Orlando RA, Kerjaschki D, Farquhar MG (1995) Megalin (gp330) possesses an antigenic epitope capable of inducing passive Heymann nephritis independent of the nephritogenic epitope in receptor-associated protein. J Am Soc Nephrol 6(1): 61-67.
- Kounnas MZ, Loukinova EB, Stefansson S, Harmony JA, Brewe BH, et al. (1995) Identification of glycoprotein 330 as an endocytic receptor for apolipoprotein J/clusterin. J Biol Chem 270(22): 13070-13075.
- Kabut J, Kondera-Anasz Z, Sikora J, Mielczarek-Palacz A (2007) Levels of complement components iC3b, C3c, C4, and SC5b-9 in peritoneal fluid and serum of infertile women with endometriosis. Fertil Steril 88(5): 1298-1303.
- Von zur Muhlen C, Schiffer E, Sackmann C, Zürbig P, Neudorfer I, et al. (2012) Urine Proteome Analysis Reflects Atherosclerotic Disease in an ApoE −/− Mouse Model and Allows the Discovery of New Candidate Biomarkers in Mouse and Human Atherosclerosis. Mol Cell Proteomics 11(7).
- Mormile R, Vittori G (2013) Endometriosis: a history written by aberrant hoxa10 gene expression and epidermal growth factor (EGF) system polymorphism? J Reprod Infertil 14(1): 46-47.
- Ejskjaer K, Sorensen BS, Poulsen SS, Mogensen O, Forman A, et al (2009) Expression of the Epidermal Growth Factor System in Eutopic Endometrium from Women with Endometriosis Differs from That in Endometrium from Healthy Women. Gynecol Obstet Invest 67(2): 118-126.
- Aghajanova L, Giudice LC (2011) Molecular Evidence for Differences in Endometrium in Severe Versus Mild Endometriosis. Reprod Sci 18(3): 229-251.
- Viganò P, Ottolina J, Sarais V, Rebonato G, Somigliana E (2017) Coagulation Status in Women With Endometriosis. Reprod Sci 25: 559-565.
- Paradisi R, Ferrini G, Matteucci C, Facchini C, Zannoni L, et al. (2017) Does exist a correlation between endometriosis and thrombophilic disorders? A pilot study. Taiwan J Obstet Gynecol 56(3): 371-373.

 Research Article
Research Article