Abstract
Introduction: The KCNQ1 gene has been known to play a role in insulin secretion and has shown association with Type 2 diabetes mellitus (T2D). The aim of the study was to investigate the association of the KCNQ1 polymorphisms rs2237892 and rs2237895 with T2D and related clinical quantitative traits.
Methods: We genotyped two single nucleotide polymorphisms (SNPs) in KCNQ1 gene, rs2237892 and rs2237895, in 230 patients with T2D and 234 controls that had no other metabolic traits in a sample obtained from Mexican population. Genotypic and allelic frequencies were evaluated, and logistic regression was used to evaluate the association between the SNPs and T2D risk.
Results: The KCNQ1 polymorphism rs2237892 was associated with T2D (odds ratio, OR: 1.58 (95% CI (1.200-2.074, P= 0.001). Also, polymorphism rs2237892 was associated with LDL-C levels (P = 0.044) in T2D patients in comparison to the control group. No significant difference in genotypic frequencies of rs2237895 was observed between T2D patients and controls.
Conclusion: This study confirms that the KCNQ1 polymorphism rs2237892 is associated with type 2 diabetes and that it influences LDL-C levels in the Mexican population.
Keywords: Endocrinology; Diabetes Mellitus; Type 2; KCNQ1 Potassium Channel; Polymorphism; Single Nucleotide; Genetic Predisposition to Disease
Introduction
T2D is the most common form of diabetes and has become one of the greatest health problems worldwide [1]. It has been documented that the population with Hispanic ancestry has a higher incidence of T2D than Caucasian population in the US [2,3]. In Mexico, according to the updated information of the International Diabetes Federation, the T2D prevalence in the population is 15.6% and it is increasing. Around 10.6 million people have T2D, 3.45 million are undiagnosed, and 7 million have impaired glucose tolerance [4]. In fact, a sharp increase of T2D has occurred due to a higher prevalence in obesity, bad dietary habits and physical inactivity. It is noteworthy to mention that the genetic background of Mexican mestizo population is complex, as it is integrated with an indigenous component and a Caucasian one, in different proportions among several country regions [5]. In northern Mexico, the European ancestry is 40-50% and the indigenous ancestry goes from 38-43%. In southern Mexico indigenous ancestry is 60-75% and European ancestry is around 10-30% [6]. Several genetic studies have been conducted in order to elucidate the molecular basis of T2D in several populations, especially in those of Caucasian background.
In Mexicans, there have been reports in which some genes showed association with early onset T2D (MODY) and with typical T2D [7-19]. However, they are present only in a small proportion of the families under study. For this reason, there is a need to search and analyze more genes that can explain the genetic basis of T2D in the Mexican population. Several reports have been published concerning the finding of KCNQ1 as a new T2D susceptibility gene. KCNQ1 encodes for a voltage-gated potassium channel expressed in pancreatic beta cells and some of its variants have been related to a decreased insulin secretion. KCNQ1 expression has also been found in cardiac muscle, intestine, and kidney. There is association of several KCNQ1 SNPs in populations in Japan, Singapore, Denmark, China, Korea, Germany, Malasya, Sweden, Finland and even Mexico [20-29]. However, in Mexican population, no analysis was made of the KCNQ1 polymorphisms with the anthropological and biochemical traits in T2D patients. Additionally, polymorphisms in this gene have been associated to gestational diabetes (GD) in China and Korea [30-32]. Our group performed some GWAS studies in populations from Starr County, Texas, and Mexico City and reported the association of several genes to T2D in these populations, including KCNQ1 gene [33-35]. In this study we analyzed if the KCNQ1 SNPs rs2237892 and rs2237895 are associated with the susceptibility to develop T2D and with the metabolic traits related to T2D in a Mexican population.
Materials and Methods
Subjects
The study included 230 unrelated Mexican-Mestizo patients with Type 2 diabetes mellitus attending the National Health Center XXI Century of IMSS in Mexico City, ranging from 30-75 years of age, according to the American Diabetes Association criteria (ADA) [36]. To reduce the probability of including control individuals who may develop T2D in later life, a group of 234 unrelated subjects without Type 2 diabetes mellitus, with no family history of diabetes and between the ages of 35–65 years made up the control group. Absence of T2D was defined as no medical history of T2D and fasting glucose levels ≤100 mg/dl. Only individuals born in Mexico whose parents and grandparents identified themselves as Mexican- Mestizos were included. The project was approved by the Ethics Committee of the National Health Center IMSS. All participants signed an informed consent before their inclusion in the study. A complete physical examination and clinical file was compiled from every participant. Weight was measured with a standardized BAME scale, model 420 and height was obtained with a standardized stadiometer in all participants.
Waist perimeter (WP) was measured in the middle point between the iliac crest and the lower rib with a flexible measure tape. Body mass index (BMI) was calculated as kilograms per meters squared. Blood pressure (BP) was measured in the right arm by the same research nurse, after sitting for at least 5 minutes, using a standard aneroid sphygmomanometer (American Diagnostic Corp.). The Korotkoff sound V was taken as the diastolic BP. Three measurements were performed within 5 min intervals and the definite value was the average of the second and third. Blood samples were taken from all individuals after fasting for 12 hours in order to determine fasting glucose, total cholesterol, HDL, LDL and triglycerides with a ILab 320 equipment (Instrumentation Laboratory SpA, Spain). Insulin was measured by chemoluminiscence with a Immulite 2000 equipment (Euro/DPC, Llanberis, UK). Insulin resistance was calculated using HOMA-IR, as insulin (UI/l) x fasting glucose (mg/dl)/22.5.
DNA Extraction
Using peripheral blood samples from patients and controls, genomic DNA extraction was performed with the commercial kit QIAamp®DNA (QIAamp DNA Blood Midi/ Kit, Qiagen, Germany) according to the manufacturer´s instructions. Purity of the samples was evaluated by spectrophotometry and the DNA integrity in electrophoresis in agarose gels at 0.8%.
SNP Genotyping
Two SNPs in the KCNQ1 gene previously reported to be associated with T2D in several populations (rs2237892 and rs2237895) were genotyped by using Taqman probes (Applied Biosystems, Carlsbad, CA, USA) for the two SNPs. Allelic discrimination was done using the fluorogenic Taqman assay, analyzed with an automated Applied Biosystems 7900 HT Real Time equipment (Applied Biosystems, Carlsbad, CA, USA). All assays were performed in duplicate and were analyzed and determined by graphic visualization using the Sequencing Detection System (SD 1.1.1., Applied Biosystems). The concordance between duplicate genotypes was greater than 90% and for quality control a call rate of 0.99 for cases and controls was used.
Statistical Analysis
Data between cases and controls was compared using chi square or Student t test for categorical and continuous variables, respectively. Hardy Weinberg equilibrium was assessed for each SNP. We evaluated the additive model for the rs2237892 and the dominant model for the rs2237895. A bivariate logistic regression was performed to calculate OR (Odds ratio) with their respective 95% CI for each one of the possible genotypic variants. The reference variant was the homozygous of the most common variant. A multivariate analysis was included to determine potential predictors of T2D risk (age, BMI and sex). In all cases, a value of p <0.05 was considered to set the statistical significance. All analyses were done using the version 11 SPSS for Windows.
Results
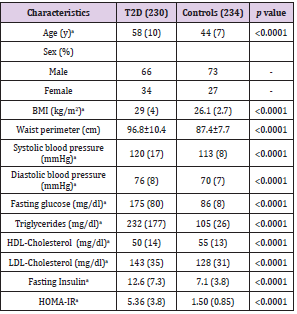
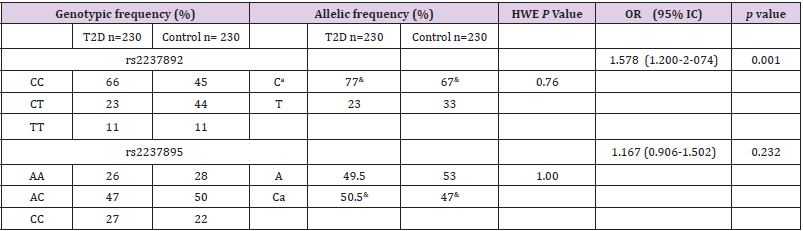
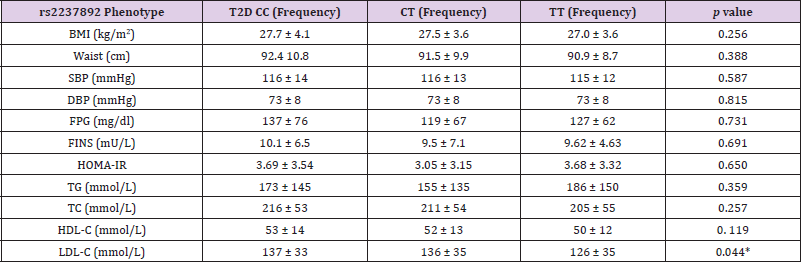
464 individuals were analyzed in this study, divided in 234 controls and 230 patients with T2D. The anthropometric and biochemical characteristics of the study population are described in Table 1. Significant differences were observed between cases and controls for the variables concerning BMI, waist perimeter, systolic and diastolic blood pressure, fasting glucose, triglycerides, HDL and LDL cholesterol, insulin and HOMA IR. Table 2 shows a descriptive analysis of the SNPs in the study, including genotypic frequencies, risk allele frequency (RAF) and Hardy Weinberg equilibrium in control individuals and patients. A greater frequency of the CC allele was observed between cases (66%) than controls (45%), although, the values turned out to be not significantly different. We did not find significant deviations from Hardy Weinberg equilibrium among controls in any of the genotyped SNPs. There were no significant differences of the genotype frequencies between the T2D group and control group without T2D or among the RAFs in these Mexican populations in comparison to the international data reported to date. In contrast, the RAF for rs2237892 in T2D patients (0.77) was significantly different to that of the control group (p= 0.001). However, the RAF in our Mexican population from both SNPs (rs2237892 and rs2237895) is slightly different when compared to RAFs reported in other populations, which shows the genetic distinctive feature of Mexican mestizos. Compared to controls, we found association between the rs2237892 polymorphism and T2D with an OR of 1.58 for the dominant model (95% CI 1.200- 2.074) estimated through conditional logistic regression (Table 2). Additionally, metabolic traits were evaluated with both SNPs and rs2237892 showed a significant association with LDL-C increase (p= 0.044) through the additive model (Table 3). There were no associations of the rs2237895 with the metabolic traits analyzed.
Table 1: General characteristics of the study population.
Note: Data reported in mean and standard deviation.
Table 2: Allelic and genotypic frequencies, Hardy-Weinberg equilibrium and association of KCNQ1 SNPs with Type 2 Diabetes in patients. (conditional logistic regression corrected by BMI, age and gender).
Note: HWE= Hardy-Weinberg Equilibrium with 2 test. a Risk allele. &= Risk allele frequency.
Table 3: Correlation analysis of metabolic traits with the genotypes of rs2237892 in T2D and control subjects.
Note: BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; FPG: fasting plasma glucose; FINS: fasting insulin; HOMA-IR: homeostatic model assessment; TG: triglycerides; TC: total cholesterol; HDL-C: high density lipoprotein cholesterol; LDL-C: low density lipoprotein cholesterol.
* Statically significant.
Discussion
The molecular mechanism in which the KCNQ1 channel is related to the pathogenesis of T2D has not been fully elucidated. However, this channel has been localized in pancreatic islets and the selective blockade of this K+ channel stimulates insulin secretion [37]. Association of the KCNQ1 gene to susceptibility to develop T2D has been reported in several GWAS studies. In fact, the two KCNQ1 SNPs analyzed in the present work (rs2237892 and rs2237895) have been found to be associated with T2D in several Asian [20-26] and European populations. These polymorphisms have also been associated to gestational diabetes (GD) in Asian populations [32,36,37]. Additionally, some KCNQ1 allelic variants (rs2237892 and rs2237895 inclusive) have associations to decreased insulin secretion, increased fasting plasma glucose and reduction of function of the beta pancreatic cells [24-31]. In this study, we observed a 0.77 RAF (C) for rs2237892 in T2D patients, which was significantly different in comparison to that in the control group (p= 0.001). Concerning the rs2237892 RAF, our Mexican population rendered the second highest value (0.95), only below that of Asian-Indians and European.
Also, we found the association of the rs2237892 SNP to T2D (OR= 1.58, p=0.001) in Mexican population through the additive model. This is interesting, as this is the highest OR score reported for this polymorphism in relation to T2D, as other OR scores range from 1.53 in China to 1.14 in several countries as Sweden, Finland, Japan, Asian and European. The OR score obtained with this Mexican population is even higher than the most recently reported for Mexican mestizo (OR= 1.37, p=0.001) [19]. This latter value was obtained using the additive model with adjustments for ancestry. The OR without ancestry adjustment in this population was weaker (OR= 1.18, p=0.062). Reasons for the differences among the OR values in the previous report and the one obtained in this study could be the inclusion criteria for the selection of T2D patients, whereas the study by Gamboa-Melendez included individuals with early onset of diabetes, as well as obese and non-obese T2D patients; in our study, all patients were confirmed for typical adult onset T2D, were non hypertensive, obese or with overweight (BMI= 29±4).
Also, the subjects in the control group were included in the study only after they were confirmed for not having any of the metabolic traits that are considered to be diagnostic for metabolic syndrome or T2D. According to this, our data confirms the previous reported association of the SNP to T2D. We further analyzed the association that these genetic variants could have with metabolic traits. The rs2237892 polymorphism was found to be associated to increased levels of L-HDL (p= 0.044) in T2D patients in comparison to controls (Table 2). In this case, a multivariated adjusted additive model was employed. There are only two previous reports on the analysis of KCNQ1 polymorphisms in relation to lipid parameters. van Vliet-Ostaptchouk et al. describes the association of the risk C-allele of rs2237892 with higher LDL-C and total cholesterol levels in T2D patients (P= 0.015 and 0.003, respectively Chen Z et al. [30] reports the association of the genotypes CC at KCNQ1 polymorphisms rs2283228 and TT at rs2237892 with higher levels of TG (P= 0.007 and 0.026, respectively) in coronary artery disease (CAD) [37].
Also, subjects with the CC genotype at rs2283228 had lower levels of HDL-C (P = 0.052). In this case, our results agree with those reported by vanVliet-Ostapchouk regarding LDL-C, although we did not see higher values of cholesterol associated with the SNPS we studied. Concerning the results by Chen Z, they are different from the values of TG and HDL-C, we observed; however, they studied patients with CAD and excluded patients with diabetes and metabolic syndrome. Even though CAD and T2D are different disorders, we realize that KCNQ1 polymorphisms show association with disturbances in lipid parameters. It is also important to remark that individuals in the control group were included in the study group only if they had normal values of BMI, blood pressure and all metabolic traits studied. This condition stresses the importance of the results obtained in this work. For rs2237895, no association with T2D was found (OR= 0.887, p=0.341). Nevertheless, this SNP RAF in Mexicans happened to be the highest of all (0.50), when compared to those reported in several other populations [31-38].
It is worth mentioning, that in other populations different KCNQ1 genotypes were associated with a reduction in insulin secretion, nonetheless, we did not find such association with fasting insulin levels. In conclusion, our data reveals the association of rs2237892 to T2D in a Mexican population, which confirms the association of some KCNQ1 genetic variants to the T2D susceptibility already reported in other populations in the world. This constitutes the first report concerning the KCNQ1 gene in Mexican mestizos and even in any Hispanic population. This is very relevant, as T2D is extremely frequent in Mexico, where the general frequency of T2D is 9.2%, although it can be as high as 19.2% in the age group of 50-59-yearolds and 25% in 60-79-year-olds. These high frequencies denote the high susceptibility that Mexicans have towards T2D, from which the genetic component should have an important role. In general, the risk allele frequencies of both rs2237892 and rs2237895 are among the highest of all RAFs reported in other populations that range from Caucasian from Europe as well as several Asian ones. It is worth mentioning that Mexican mestizos arose from the mixture from Amerindian and Caucasian in varying proportions. To be able to have a better understanding of these SNPs RAFs nature, confirmation of the association between these alleles in indigenous Mexican populations is yet to be obtained.
Acknowledgement
This work was supported by the National Polytechnic Institute for the performance of the studies mentioned in the article as well as by CEDOPEC. The authors declare no conflict of interest.
References
- (2011) Centers for Disease Control and Prevention. National diabetes fact sheet: national estimates and general information on diabetes and prediabetes in the United States, 2011. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention.
- Harris MI, Flegal KM, Cowie CC, Eberhardt MS, Goldstein DE, et al. (1998) Prevalence of diabetes, impaired fasting glucose, and impaired glucose tolerance in U.S. adults: The Third National Health and Nutrition Examination Survey, 1988-1994. Diabetes Care 21(4): 518-524.
- Harris MI (1998) Diabetes in America: epidemiology and scope of the problem. Diabetes Care 21(3):C11-4.
- (2011) International Diabetes Federation. IDF Diabetes Atlas, 5th edn. Brussels, Belgium: International Diabetes Federation.
- Barquera Cervera S, Rivera Dommarco J, Campos Nonato I, Hernández Barrera L, Santos Burgoa Zarnecki C, et al. (2010) Acuerdo Nacional para la Salud Alimentaria. Estrategia contra el sobrepeso y la Obesidad. Secretaria de Salud.
- Rangel Villalobos H, Muñoz Valle JF, González Martín A, Gorostiza A, Magaña MT, et al. (2008) Genetic admixture, relatedness, and structure patterns among Mexican populations revealed by the Y-chromosome. Am. J. Phys. Anthropol 135: 448-461.
- Domínguez López A, Miliar García A, Segura Kato YX, Riba L, Esparza López R, Ramírez Jiménez S, et al. (2005) Mutations in MODY genes are not common cause of early-onset type 2 diabetes in Mexican families. JOP 6(3): 238-245.
- Villarreal Molina MT, Flores Dorantes MT, Arellano Campos O, Villalobos Comparan M, Rodríguez Cruz M, et al. (2008) Association of the ATP-binding cassette transporter A1 R230C variant with early-onset type 2 diabetes in a Mexican population. Metabolic Study Group Diabetes 57(2): 509-513.
- Menjívar M, Granados Silvestre MA, Montúfar Robles I, Herrera M, Tusié Luna MT, et al. (2008) High frequency of T130I mutation of HNF4A gene in Mexican patients with early-onset type 2 diabetes. Clin. Genet 73(2): 185-187.
- Cruz M, Montoya C, Gutiérrez M, Wacher NH, Kumate J, et al. (2002) Polimorfismos de genes relacionados con Diabetes tipo 2. Rev Med IMSS 40: 113-125.
- Del Bosque Plata L, Aguilar Salinas CA, Tusié Luna MT, Ramírez Jiménez S, Rodríguez Torres M, et al. (2004) Association of the calpain-10 gene with type 2 diabetes mellitus in a Mexican population. Mol Genet Metab 81(2): 122-126.
- Parra EJ, Cameron E, Simmonds L, Valladares A, McKeigue P, et al. (2007) Association of TCF7L2 polymorphisms with type 2 diabetes in Mexico City. Clin Genet 71(4): 359-366.
- Burguete Garcia AI, Cruz Lopez M, Madrid Marina V, Lopez Ridaura R, Hernández Avila M, et al. (2010) Association of Gly972Arg polymorphism of IRS1 gene with type 2 diabetes mellitus in lean participants of a national health survey in Mexico: a candidate gene study. Metabolism 59(1): 38-45.
- Cruz M, Valladares Salgado A, Garcia Mena J, Ross K, Edwards M, et al. (2010) Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev 26(4): 261-270.
- Martínez Gómez LE, Cruz M, Martínez Nava GA, Madrid Marina V, Parra E, et al. (2011) A replication study of the IRS1, CAPN10, TCF7L2, and PPARG gene polymorphisms associated with type 2 diabetes in two different populations of Mexico. Ann Hum Genet 75(5): 612-620.
- Guzmán Flores JM, Muñoz Valle JF, Sánchez Corona J, Cobián JG, Medina Carrillo L, et al. (2011) Tumor necrosis factor-alpha gene promoter -308G/A and -238G/A polymorphisms in Mexican patients with type 2 diabetes mellitus. Dis Markers 30: 19-24.
- Gutiérrez Vidal R, Rodríguez Trejo A, Canizales Quinteros S, Herrera Cornejo M, Granados Silvestre MA, et al. (2011) LOC387761 polymorphism is associated with type 2 diabetes in the Mexican population. Genet. Test Mol Biomarkers 15(1): 79-83.
- Martínez Calleja A, Quiróz Vargas I, Parra Rojas I, Muñoz Valle JF, Leyva Vázquez MA, et al. (2012) Haplotypes in the CRP gene associated with increased BMI and levels of CRP in subjects with type 2 diabetes or obesity from Southwestern Mexico. Exp Diabetes Res 2012: 982683.
- Gamboa Meléndez MA, Huerta Chagoya A, Moreno Macías H, Vázquez Cárdenas P, Ordóñez Sánchez ML, et al. (2012) Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes 61(12): 3314-3321.
- Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, et al. (2008) SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet 40(9): 1098-10102.
- Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, et al. (2008) Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 40(9): 1092-1097.
- Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, et al. (2008) Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet 53(11): 991-998.
- Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, et al. (2009) Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia 52(7): 1315-1321.
- Hu C, Wang C, Zhang R, Ma X, Wang J, et al. (2009) Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia 52(7): 1322-1325.
- Qi Q, Li H, Loos RJ, Liu C, Wu Y, et al. (2009) Common variants in KCNQ1 are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Hum Mol Genet 18(18): 3508-3515.
- Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, et al. (2009) Genetic variation in KCNQ1 associates with fasting glucose and beta-cell function: a study of 3734 subjects comprising three ethnicities living in Singapore. Diabetes 58(6): 1445-1449.
- Holmkvist J, Banasik K, Andersen G, Unoki H, Jensen TS, et al. (2009) The Type 2 Diabetes Associated Minor Allele of rs2237895 KCNQ1 Associates with Reduced Insulin Release Following an Oral Glucose Load. PLoS ONE 4: 5872.
- Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, et al. (2009) A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes 58(10): 2409-2413.
- Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, et al. (2009) Association of type 2 diabetes candidate polymorphisms in KCNQ1with incretin and insulin secretion. Diabetes 58(7): 1715-1720.
- Van Vliet Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, et al. (2012) Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS ONE 7(3): e32148.
- Boini KM, Graf D, Hennige AM, Koka S, Kempe DS, et al. (2009) Enhanced insulin sensitivity of gene targeted mice lacking functional KCNQ1. Am J Physiol Regul Integr Comp Physiol 296(6): 1695-1701.
- Kwak SH, Kim TH, Cho YM, Choi SH, Jang HC, et al. (2010) Polymorphisms in KCNQ1 are associated with gestational diabetes in a Korean population. Horm Res Paediatr 74(5): 333-338.
- Cruz M, Valladares Salgado A, Garcia Mena J, Ross K, Edwards M, et al. (2010) Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev 26(4): 261-270.
- Parra EJ, Below JE, Krithika S, Valladares A, Barta JL, et al. (2011) Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican American sample from Starr County, Texas. Diabetologia 54(8): 2038-2046.
- Below JE, Gamazon ER, Morrison JV, Konkashbaev A, Pluzhnikov A, et al. (2011) Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia 54(8): 2047-2055.
- Shin HD, Park BL, Shin HJ, Kim JY, Park S, et al. (2010) Association of KCNQ1 polymorphisms with the gestational diabetes mellitus in Korean women. J Clin Endocrinol Metab 95(1): 445-449.
- Zhou Q, Zhang K, Li W, Liu JT, Hong J, et al. (2009) Association of KCNQ1 gene polymorphism with gestational diabetes mellitus in a Chinese population. Diabetologia 52(11): 2466-2468.
- Chen Z, Yin Q, Ma G, Qian Q (2010) KCNQ1 gene polymorphisms are associated with lipid parameters in a Chinese Han population. Cardiovasc. Diabetol 9: 35.

 Research Article
Research Article