Abstract
PCR arrays are designed to produce gene expression profiles of a subset of genes associated with biological processes, signal transduction mechanisms, and diseases. The current review addresses how PCR arrays can be used to explore human immunity. Human immune responses are divided into two major phases: innate immunity and adaptive immunity. Innate immunity consists of host puerperal defenses that protect the human body from foreign invaders using anatomical, chemical, and nonspecific cellular sentries. Adaptive immunity consists of pathogen-specific defenses that utilize B cells and T cells to detect, target, and eliminate pathogens and tumor cells from the human body. Recent research studies using PCR arrays have enhanced our understanding of immune mechanisms in the context of infectious diseases, however, more work needs to be done in this area. Gene expression profiles associated with human immunity may lead to the design of therapeutic interventions to attenuate pathophysiological outcomes associated with inflammation or treat microbial diseases. PCR array gene expression profiles may also lead to the identification of either up-regulated genes or down-regulated genes that produce favorable immune responses and may serve as novel biomarkers. The article also discusses PCR array experimental designs and lists commercial PCR arrays that focus on elucidating new insights into innate and adaptive immunity.
Introduction
Gene expression is a complex process whereby genes produced via transcriptional processes in the nucleus for eukaryotes and the cytoplasm for prokaryotes are converted into a functional protein via the process of translation in the cytoplasm in living organisms. Gene expression is very important to many researchers from biology and biology-related disciplines. Prior to the invention of the PCR machine [1], gene expression studies were primarily performed using Northern blots and Western blots. Real-time PCR machines have opened the door to PCR arrays, a sensitive high-throughput technique for monitoring gene expression. Human immunity is an important biological process that protects humans against pathogens and cancer [2,3]. Human immunity is largely broken down into two major groups: innate immunity and adaptive immunity. Innate immunity primarily consists of anatomical barriers, antimicrobial substances, and immune cells that are designed to respond to any pathogen. Adaptive immunity consists of B cells and T cells and is designed to target a specific pathogen. In general, innate immunity is designed to keep pathogens outside the human body and to quickly eliminate them if they breach the internal environment, while adaptive immunity or acquired immunity is designed to neutralize and eliminate a specific pathogen that has evaded the innate immune surveillance system [4]. The human immune system is armed with extremely powerful weapons that help promote a homeostatic environment. Since the days of Edward Jenner’s cowpox experiments where the basic features of immunity were established, immunological investigations have produced remarkable research findings and greatly expanded our current understanding of the complexity of the human immune system [4,5].
However, with all biological systems a complete understanding of paramount molecular interactions, signal transduction mechanisms, and molecular physiology will require new experimental technologies as well as sensitive instrumentation to elucidate more details regarding human immunity. Immune responses are the result of thousands of unique molecular mechanisms that coordinate hematopoietic cells, cytokines, hormones, and growth factors. Understanding the precise molecular network responsible for innate and adaptive immunity will help immunologists and pharmaceutical scientists design strategies for dealing with cancer and infectious diseases. A very powerful weapon in the pursuit of knowledge of human immunity are PCR arrays.
PCR arrays are designed to gain insight into specific biological mechanisms and genetic function. Unlike microarrays which have the capacity to monitor thousands of genes at one time, PCR arrays typically monitor less than 100 genes. Microarrays focus on global gene expression, while PCR arrays are constructed to monitor specific cells, biologic processes, diseases, and normal and abnormal human physiology. The advantages to using a PCR array are that they are far less expensive to perform than a traditional microarray experiment. Both PCR arrays and microarrays involve similar sample preparation protocols such as the growth of target cells and tissue using standard tissue culture protocols.
PCR Arrays
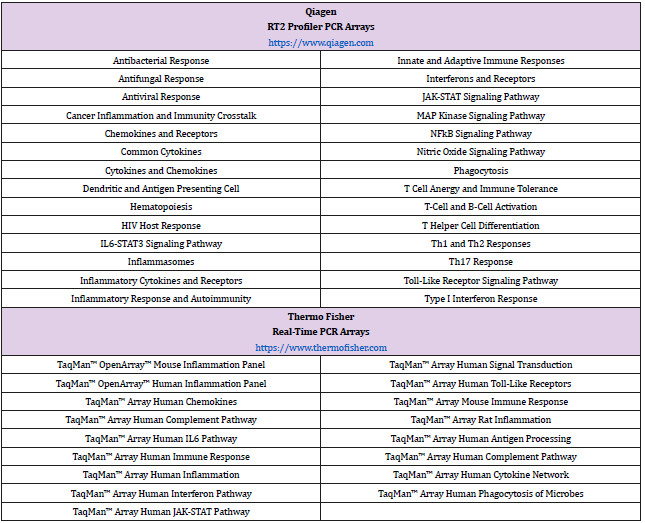
PCR arrays are designed by companies who engage in the meticulous culling of hundreds of research articles and review papers that identify key genes involved with human immune responses. The bibliographic data is then used to create a particular PCR array consisting of research-relevant genes. Specific primers of identified genes are then immobilized onto a 96-well plate or 384- well plate. Prior to the addition of complementary DNA (cDNA) from the experimental cellular system, cDNA is mixed with a master mix that typically contains SYBR green or similar molecule that will aid in the detection of amplified products during the data acquisition phase. There are a number of innate and adaptive immunity-based PCR arrays on the market (Table 1) lists a sample of the PCR arrays that immunologists and microbiologists may find useful to explore both innate and adaptive immunity.
PCR Array Protocols to Explore Immunity
Host-pathogen interactions involve the highly coordinated sequence of molecular events mediated by the pathogen and host organism. Pathogens utilize a molecular arsenal to gain entry into the host tissues and cellular environments. Microbial inhibitory molecules such as interferons, interleukins, complement, and other cytokines and are recruited to block entry of microorganisms. In terms of tumor infiltration, tumor cells secrete a wide array of molecules to support tumor growth and metastasis to ectopic regions of the human body. During these events the immune system is also busy producing anti-tumor substances to control cell proliferation and metastasis [6]. Not surprisingly, an extraordinarily complex picture begins to emerge when you cogitate the molecular interplay between tumor cells and the human immune system.
PCR arrays are particularly well suited to enhance our understanding of the human immune response. Specifically, PCR arrays could be used to identify a protein or family of proteins that play a major role during disease progression or elucidate biomarkers that may provide a surveillance system during infection or cancer development. The data interpretation phase in PCR array studies is equally as critical as the data analysis phase. A hostspecific down-regulated gene detected via bioinformatics analysis could imply that the host is attempting to modify a particular biological structure or signal transduction pathway that is beneficial to the pathogen. Alternatively, a down-regulated host gene could indicate a pathogen-mediated approach to minimize the effects of an antimicrobial molecule. Thus, investigators must perform additional experiments to determine the cause of a particular PCR array-derived phenomenon.
The key to designing a PCR array experiment that conveys meaningful insight regarding innate and adaptive immunity is the inclusion of both biological replicates and technical replicates. Biological replicates are designed to investigate variation that exists due to the biological phenomenon under study. Biological replicates involve performing the same experimental procedure on at least two or more different biological samples. Ideally, researchers should generate similar biological outcomes on each biological replicate. If one sample shows a significant difference compared to the other samples then the results may not be generalizable and may be due to random biological events. Since the samples in a biological replicate are distinctly different samples from different animals, separate tissue culture flasks, or separate cell stocks similar results from each sample provides strong evidence that the biological effect is not random and generalizable to the cellular or organismal system.
Technical replicates are designed to determine if variation exists due to the technique or experimental procedure employed. Technical replicates involve performing the same procedure multiple times (e.g., two or more) using the same biological sample. Technical replicates address issues such as reproducibility of the protocol or variation in the data due to the technique or assay procedures. Ideally, researchers expect to produce similar results for each technical replicate. Technical replicates generate either confidence or concern in the selected experimental approach or equipment and must be added to the experimental design. Since the samples of the replicate series are the same, each test should yield the same result. For PCR array experiments, an example of a technical replicate would be to add three aliquots of cDNA from the same sample to three different PCR array wells. A sufficient biological replicate would be to isolate RNA and convert RNA to cDNA using three different culture flaks of the same cell line. Both technical replicates and biological replicates enhance the experimental design and improve the overall statistical power.
Immunologists, microbiologists, and cancer researchers have a wide range of experimental design options when planning PCR array experiments to explore immunity. To ascertain gene expression changes from newly discovered cytokines, an immunologist could treat various hematopoietic cells with cytokines and add cellular cDNA to an appropriate PCR array. The effects of paracrine and autocrine signaling mechanisms could also be explored using this approach. Drug and pharmaceutical combination studies could be performed to determine the precise drug combinations required to activate the immune system. Every known pathogen could be tested on specific cell types to study innate and adaptive immunity ramifications. The ability of a microbial pathogen to activate host immune gene expression could indicate unknown pathogen-specific evasion mechanisms and may suggest beneficial antimicrobial strategies for certain susceptible cells.
The current and rapidly growing selection of PCR arrays also allow investigators the opportunity to determine the impact of pathogenic infections on specific signal transduction mechanisms. Disease-specific PCR arrays could allow scientists to determine the extent to which intracellular pathogens up-regulate or downregulate disease biomarkers. Both primary cancer cells and immortal cancer cell lines can be studied using immunity-based PCR arrays to further explore immunity mechanisms associated with tumor development, tumor progression, and tumor metastasis. Moreover, dose-dependent PCR array-based studies could be performed to determine the efficacy of therapeutic compounds to produce a pro-immunity gene expression profile. Time-dependent studies involving therapeutic compounds could also provide insight into temporal gene expression changes beneficial to human health. Time-course PCR array studies tracking various stages of a particular pathogenic infection in human cells would provide invaluable data regarding gene expression patterns that support innate and adaptive immune mechanisms.
Inflammation has been shown to play a major role in pathophysiological outcomes of many diseases [7-9]. Inflammation is an innate immune response to any type of tissue damage whether the source is endogenous (e.g., cancer) or exogenous (e.g., pathogen). Inflammation occurs in several stages and involves recruitment of blood cells, phagocytes, and chemical factors designed to neutralize the threat and aid in the process of wound repair. An understanding of pro-inflammatory and anti-inflammatory gene expression changes that occur during the course of an infection or during various stages of disease progression may contain beneficial evidence regarding potentially effective therapeutic treatments. Treatment studies such as the use of monoclonal antibodies to block or neutralize specific inflammatory-associated proteins may suppress tissue damage associated with inflammation [10]. The synthesis of transcription and translation inhibiting molecules that target deleterious inflammatory proteins and genes may also prove beneficial.
Silberstein et al. [11] demonstrated the utility of PCR arrays to investigate the role of apoptosis in liver damage following chronic hepatitis C virus (HCV) infection. The Human Apoptosis RT2 Profiler PCR Array from Qiagen was used to examine cDNA from cells infected with HCV. Data analysis indicated that major apoptotic genes such as tumor necrosis factor, caspases, and Bcl2- interacting killer were involved in liver cell death induced by HCV. Interestingly Han et al. [12] performed a qPCR array experiment on rainbow trout expressing the cecropin P1 transgene. Cecropin is a protein isolated from the porcine digestive tract that has broad spectrum antibacterial activity [13] and antiviral activity [14]. PCR primers that are unique to innate and adaptive immune mechanisms were used in the PCR array assay to demonstrate the immune related genes that play a role in the antimicrobial effects of cecropin in rainbow trout. Data revealed that the antimicrobial effects were mediated in part due to cytokine and chemokine signaling, complement genes, antigen processing and presentation, lysosomal phagocytosis, and toll-like receptor signaling
Conclusion
PCR arrays utilize real-time PCR technology to identify gene expression patterns and molecular networks associated with biologic phenomena. PCR arrays can be more effective at resolving molecular events than microarrays due to the small subset of genes present on each array plate. Further, because each PCR array focuses on a specific biological pathway, researchers can pinpoint which genes contribute to normal and aberrant physiologic processes. As with any biological investigation, the design of your experiment is paramount. The inclusion of the proper biological replicates and technical replicates will facilitate data analysis [15,16].
While there are a number of published reports that highlight the use of PCR arrays to explore host-pathogen interactions, more studies are needed to construct a more complete picture of how the human immune system responds to infection and cancer. Using PCR arrays will undoubtedly shed light on previously unknown molecules that participate in crosstalk mechanisms and serve as molecular adaptors critical in establishing a strong defense system against internal and external threats. Since PCR arrays work using fluorescence detection, real-time PCR machines are required. To facilitate data analysis, most PCR array manufacturers offer free or inexpensive bioinformatics software.
Acknowledgement
This work was supported by grants funded by the National Science Foundation (HRD-1533536 and HRD-1505098).
Conflict of Interest
No conflict of interest.
References
- Shampo M, Kyle R (2002) Kary B. Mullis-Nobel laureate for procedure to replicate DNA. Mayo Clinic Proceedings 77(7): 606.
- Sima P, Vannucci L, Vetvicka V (2019) Immunity in cancer and atherosclerosis. Annals of Translational Medicine 7(9): 1-5.
- Thakur A, Mikkelsen H, Jungersen G (2019) Intracellular pathogens: Host immunity and microbial persistence strategies. Journal of Immunology Research 1-24.
- Sattler S (2017) The role of the immune system beyond the fight against infection. Advances in Experimental Medicine and Biology 1003: 3-14.
- Brodin P, Davis M (2017) Human immune system variation. Nature Reviews Immunology 17(1): 21-29.
- Croci D, Salatino M (2011) Tumor immune escape mechanisms that operate during metastasis. Current Pharmaceutical Biotechnology 12(11): 1923-1936.
- Greten F, Grivennikov S (2019) Inflammation and cancer: Triggers, mechanisms and consequences. Immunity 51(1): 27-41.
- Halim M, Halim A (2019) The effects of inflammation, aging and oxidative stress on the pathogenesis of diabetes mellitus (type 2 diabetes). Diabetes & Metabolic Syndrome 13(2): 1165-1172.
- Staudacher A, Stevens W (2019) Sinus infections, inflammation, and asthma. Immunology and Allergy Clinics of North America 39: 403-441.
- Hatano R, Itoh T, Otsuka H, Okamoto S, Komiya E, et al. (2019) Characterization of novel anti-IL-26 neutralizing monoclonal antibodies for the treatment of inflammatory diseases including psoriasis. Monoclonal Antibodies 18: 1-15.
- Silberstein E, Ulitzky L, Lima L, Cehan N, Teixeira-Carvalho A, et al. (2016) HCV-mediated apoptosis of hepatocytes in culture and viral pathogenesis. Public Library of Science One 11(6): 1-20.
- Han Y, Chen T (2019) A pathway-focused RT-qPCR array study on immune relevant genes in rainbow trout (Oncorhynchus mykiss) harboring cecropin P1 transgene. Fish and Shellfish Immunology 89: 1-11.
- Zhou Y, Peng Y (2013) Synergistic effect of clinically used antibiotics and peptide antibiotics against Gram-positive and Gram-negative bacteria. Experimental and Therapeutic Medicine 6(4): 1000-1004.
- Guo C, Huang Y, Cong P, Liu X, Chen Y, et al. (2014). Cecropin P1 inhibits porcine reproductive and respiratory syndrome virus by blocking attachment. BMC Microbiology 14: 1-11.
- Brazma A (2009) Minimum information about a microarray experiment (MIAME) successes, failures, challenges. Scientific World Journal 9: 420-423.
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics 29(4): 365-371.

 Review Article
Review Article