Abstract
The spread of antibiotic resistant bacteria is a growing problem and a public health issue. Over the years, various genetic mechanisms concerned with antibiotic resistance have been identified to be natural and acquired resistance. The natural resistance involved mutation via target modification, reduced permeability, efflux system and on the other hand, acquired resistance via horizontal gene tranfer include Moblie Genetic Elements [MGEs] such as plasmid, transposon and integrons genetic elements that can acquire, exchange, and express genes embedded within Gene Cassettes [GC]. Integrons are widely distributed, especially in Gram-negative bacteria; they are carried by Mobile Genetic Elements such as plasmids, and transposons, which promote their spread within bacterial communities and have been studied mainly in the clinical setting for their involvement in antibiotic resistance, their role in the environment is now an increasing focus of attention. The aim of this review is to educate the populise about the mechanisms of multiple drug resistance bacteria isolates and the danger ahead if appropriate regulations are not put in place especially in developing country like Nigeria.
Keywords: Antibiotics; Multiple Drug Resistant Bacteria; Mobile Genetic Element; Integrons Plasmid
Abbreviations: GC: Gene Cassettes; DNA: Desoxyribonucleic Acid; RNA: Ribonucleic Acid; WHO: World Health Organization; HGT: Horizontal Gene Transfer; LPS: Lipopolysaccharide; QRDR: Quinolone-Resistance Determining Region; MGC: Mobile Genetic Components; RIs: Resistant Integron; CIs: Chromosomal Integrons; MIs: Mobile Integrons; MRIs: Multidrug Resistance Integrons
Introduction
Microbes that are causative agents of infectious disease represent a serious public health concern globally. The invention and use of antibiotics have been one in all the main scientific achievements of the 20th century. Antibiotics are either cytotoxic or cytostatic to the microorganisms, permitting the body’s natural defenses, like the immune system, to flush them out. They often act by inhibiting the synthesis of bacterial cell, synthesis of proteins, Desoxyribonucleic Acid [DNA], Ribonucleic Acid [RNA], by a membrane disorganizing agent, or alternative specific actions [1]. The primary antibiotic, penicillin, was discovered in 1928 by Sir Alexander Fleming, a Scottish biologist and pharmacologist. Antibiotics could enter the semipermeable membrane of the microorganism and bind to them, using the energy-dependent transport mechanisms in ribosomal sites, which then cause the inhibition of the macromolecule synthesis [1]. In the struggle against infections or microorganisms, beyond any doubt antibiotics are a blessing to human civilization that has saved many individuals [2]. Completely different antibiotics are being used for therapeutic purpose over time. Antibiotics were seen as the ‘wonder drug’ within the mid-20th century [3]; and were thought of as cure that by selection targeted microbes that were chargeable for disease causation, however without having side effect on the host. In the era from the 1950s to 1970s was therefore seen as the golden era for the development of novel antibiotics categories [4]. Throughout the previous few decades, incidence of microorganism infections has improved dramatically. Continuous exploitation of antimicrobial medicine in the cure of infections has resulted in the emergence of resistance among pathogens as well as multiple drug resistant strains [5].
Moreso, the haphazard and ridiculous use of antibiotics today has led to the evolution of recent resistant strains of microorganism that are somewhat deadly compared to the parent strain. Cases of widespread prevalence of resistant microorganism are currently quite common that ends up in several health-related issues [6]. Modification within the genetic formation of these resistant microorganisms is so quick that the effectiveness of common antibiotics could be lost within few years from now [7]. As per the report of the World Health Organization [WHO], resistance in most of the deadly infectious microorganism infections worldwide like respiratory tract infections, diarrhea, meningitis, syphilis, venereal disease and tuberculosis [8]. The aim of this review is to coach the populise concerning the event and mechanisms of multiple drug resistance bacterial isolates and also the danger ahead if applicable laws are not put in place particularly in developing African nation like Nigeria.
Development of Antibiotic Resistance
Bacterial battle to become resistance to antibiotic tends to possess a natural process that encourages resistance. This happens as a natural organic process response of bacteria following their exposure to these antimicrobials and is defined as the capability of microorganism to survive the repressing concentration of an antibiotic. It may be natural [intrinsic] or acquired [noninheritable]. The former is the resistance characteristic for all members of a given microorganism species or genus. It results from the inability of the antibiotic to succeed in its target, lack of affinity for the target, the presence of outflow pumps or possession of alternative body resistance mechanisms [9]. However, noninheritable resistance is much more necessary and vital owing to the potential spread of resistant genes through a sensitive microorganism population. These genes may be collected on a range of gene transfer systems, like bacterial conjugative plasmids, transposable elements and integron systems, that move the genes answerable for antibiotic resistance from one DNA system to a different system and from a resistant microorganism to a sensitive one [10]. Thus, selective stress phenomena are thought to occur in area wherever antimicrobial agents are broadly used, like in human clinical medicine [11], agriculture, in natural soil and aquatic environments [12].
Multiple Drug Resistance
The development of Multiple Drug Resistant [MDR] microbes arises from the utilization of various antibiotics available. MDR is that the ability of microbes to show resistance against three or a lot of categories of antibiotics inhibition concentration. It is additionally seen as the insensitivity of microbes to the administered antimicrobial medicines despite earlier sensitivity to that same drug [13]. Microorganisms could collect multiple resistance traits over time and might become immune to multiple categories of antibiotics [14]. Microorganisms get mutated/adapted to reduce or eliminate the potency of antibiotic and still survive while not damage. According to World Health Organization, these resistant microorganisms are able to contest attack by antimicrobial medicine, which ends up in unsuccessful treatment leading to persistence and spreading of infections. Therefore, antimicrobial use fosters microorganism drug resistance and dissemination of drug resistance determinants at intervals populations. Multidrug resistant microorganism could also be recalcitrant to clinically relevant therapeutic agents, leading to treatment failures of infectious disease [15].
Although the event of MDR may be a natural prevalence, intensive rise within the range of upset conditions, like diabetic patients, HIVinfection, persons who have undergone organ transplantation, and severe burn patients, makes the body an easy target for hospital noninheritable infectious diseases, thereby making the body conducive to more unfold of MDR. There are several research papers that have completely identified different bacteria species that have shown resistance to multiple antibiotics. A number of them are Acinetobacter baumannii [16,17]; Staphylococcus aureus [18,19]; Enterococcus species [20-22]; Clostridium difficile [23]; Klebsiella pneumoniae [24,25]; Streptococcus pneumoniae [26]; Mycobacterium tuberculosis [27]; Salmonella enterica [28]. MDR may be a threat to developing countries and also the world at large.
Consequence of Multidrug Resistance
Antibiotic resistant microbes are called superbugs. They are not solely a laboratory concern but have however become a world threat accountable for high death tolls and life-threatening infections [29]. Antimicrobials have been used for several years across the world till date. Shut watch in numerous regions of the planet as well as Africa has shown that a lot of infectious microorganisms have evolved over the years and there is a fearsome threat of high range of antibiotic-resistant species which can resist the repressing effects of these antibiotics. Multidrug resistance is one of the reasons why microbes fail to respond to typical medicine, thus, extending the length or course of treatment more and therefore increasing the health care cost which tend to worsen position of individuals who are not capable of such expenses. The probability of controlling several infectious diseases have reduced due to resistance to relevant antibiotics, therefore making it a global concern.
Problems Related to Multidrug Resistance
Resistance to antibiotics is linked with high death rates; high medical cost and has a momentous blow on the effectiveness of antimicrobial agents. MDR act as a barrier in disease management by increasing the chance of spreading of resistant pathogens, thereby declining effectiveness of treatment and, thus, leading to prolonged time of infection in patient. The cost of treatment is increased due to MDR as the pathogens became resistant to commercially accessible medicine, which has triggered the utilization of more expensive therapies [30]. The pace of success of present-day medical applications like cancer therapy and organ transplantation has contributed vastly towards development of MDR. Variations within the resistance profiles of pathogens as well as the quality of public hygiene also have a substantial impact on the effectiveness of antimicrobial agents. Expansion of global trade and tourism lead to increased potential of MDR to spread all over the world and decrease in export and import of various products affecting the economy of developing countries [31].
Molecular Mechanism of Microorganism Resistance to Antibiotics
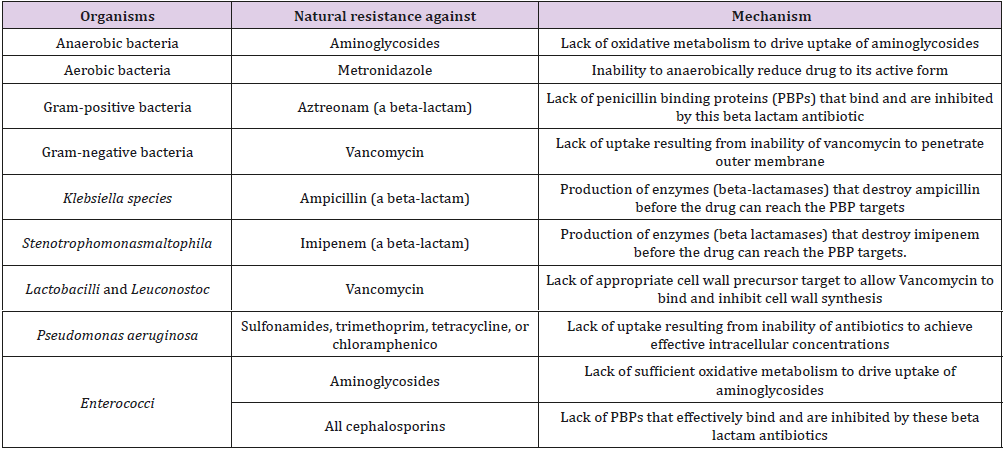
In quest to grasp the molecular mechanisms essential for antibiotic resistance, understanding of microorganism structure and performance is core [32]. Though many new drugs have been introduced commercially, the event of resistance among infectious microorganisms is increasing particularly in patients below prolonged drug exposure [33]. Molecular mechanism of microorganism antibiotics resistance is genetically divided into two categories: natural [intrinsic] resistance and acquired resistance. Intrinsic resistance refers to the condition where bacterial species is not affected by antibiotic due to its fundamental physiological properties like Vancomycin resistance in Enterobacteriaceae due to the outer membrane of Gram-negative species. In contrast, noninheritable resistance refers to the situation whereby a bacterium that use be sensitive to antibiotic at a given concentration is not any longer suppressed at that very same concentration [34].
As a matter of reality, solely non-inheritable resistance has vital clinical implications. This is often so as the emergence of resistance renders formerly efficient treatments useless, particularly within the transition phase where resistance is just too low to encourage a modification within the empirical treatment. Non-inheritable resistance can be further divided into horizontally non-inheritable resistance and mutational acquired resistance. Horizontally noninheritable resistance refers to the Horizontal Gene Transfer (HGT), usually within the plasmid conjugation, phage transduction or non-specific DNA uptake. On the opposite hand, mutational noninheritable resistance occurs when the bacterial genome mutates to overcome the impact of an antibiotic and ordinarily involves the modification of one or few nucleotides. Both horizontally and mutational non-inheritable resistance play a significant role in clinically relevant resistance [35].
Intrinsic Resistance
This is the inborn ability of a microorganism to resist activity of a selected antimicrobial agent through its inherent structural or functional characteristics, which permit tolerance of a selected drug or antimicrobial category. This is often referred to as insensitivity since it happens in organisms that have not been vulnerable to that exact drug. Such natural unfitness may be due to lack of affinity of the drug for the microorganism target, unavailability of the drug into the microorganism cell, pumping out of drug by chromosomally encoded active exporters, innate production of enzymes that inactivate the drug [36]. Intrinsic mechanisms are those specified by naturally occurring gene found on host chromosome. Efflux systems in Gram-positive microorganism involve a single polypeptide within the cytoplasmic membrane. Gram negative bacterial cell envelope may be a major barrier for antibiotics and consists of the cell membrane, the periplasm and also the outer membrane. The outer membrane is the major barrier that forestalls antibiotics penetration through porins or by passive diffusion through the outer membrane phospholipids bilayer. However, poly cationic compounds like Gentamicin and colistin are being transported through the outer membrane through interaction with Lipopolysaccharide (LPS) in a method known as self-promoted uptake [37].
It has been projected that intrinsic resistance is chiefly the consequence of the solidity of cellular envelopes, the activity of multidrug outflow pumps or the shortage of applicable targets for a given family of medication. However, recently accessible articles indicate that the characteristic phenotype of susceptibility to antibiotics of a given bacterial species depends on the combined activity of many parts known as intrinsic resistome. The three most relevant causes of intrinsic resistance are: lack of the target, activity of chromosomally encoded antibiotic-inactivating enzymes and reduced uptake of the antibiotic [38] (Table 1).
Acquired Resistance
Bacteria also can acquire resistance. This happens when a type of bacteria changes in a manner that protects it from the antibiotic. Changes in bateria genome on the other hand, could consequently result in a modification within the nature of proteins expressed by the organism. Such modification can result in structural and functional alteration in the features of the bacteria involved, which may end in changes resulting in resistance against a selected antibiotic [39]. From an evolutionary perspective, bacteria use two major genetic methods to adapt to the antibiotic “attack” (i) mutations in gene[s] typically related to the mechanism of action of the compound, and (ii) acquisition of foreign DNA coding for resistance determinants through horizontal gene transfer (HGT).
Mutational Resistance
Is a spontaneous modification within the DNA sequence within the gene which may result in modification of the character that it codes for [15]. These bacterial cells are derived from a vulnerable population that developed mutations within the genes that have effect on the activity of the drug, leading to preserved cell survival within the presence of the antimicrobial molecule. This is often so because a modification in a single nucleotide could result in corresponding modification in one or more amino acid which can then change the enzyme or cell structure that consequently changes the affinity or effective activity of the targeted antimicrobials. Once a resistant mutant emerges, the antibiotic eliminates the susceptible population and as such, the resistant microorganism predominates. In general, mutations leading to antimicrobial resistance alter the antibiotic action via one of the following mechanisms,
(i) Modifications of the antimicrobial target (decreasing the affinity for the drug),
(ii) Reduced perm ableness,
(iii) Activation of efflux mechanisms to squeeze out the harmful molecule, or
(iv) Global changes in necessary metabolic pathways via modulation of regulative networks [40].
Target Modification
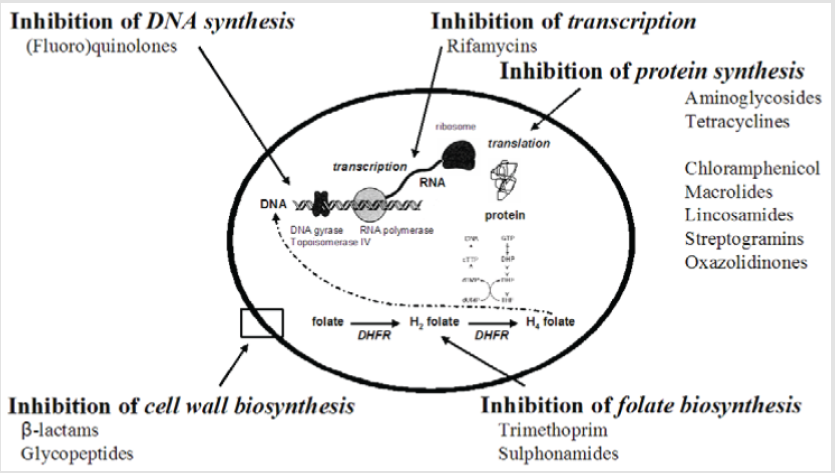
Bacteria have found ways to change the molecular targets of antimicrobial agents. Natural variations or non-inheritable changes within the target sites of antimicrobials that forestall drug binding or action may be a common mechanism of resistance. Target site changes; typically result from spontaneous mutation of a bacterial gene on the chromosome and selection in the presence of the antimicrobial. This sort of resistance mechanism is shown by both Gram-negative as well as Gram-positive. The mechanism of action for many of the antibacterial drug involves interaction between the drug and intracellular enzyme/protein. The development of drug resistance for these antibiotics requires the reduction in affinities to their enzymatic targets. Altering antibiotic’s target super molecule directly at the DNA level may be a common mechanism of target modification [41]. Altered targets could embody, as an example, DNA gyrase, a target of quinolone antimicrobials [42], RNA polymerase, a target of rifampin [43], prokaryotic ribosome, a target of tetracycline and other protein synthesis inhibitors [44].
Another example of altered target mechanism includes substitution of amino acids within the Quinolone-Resistance Determining Region [QRDR] of DNA gyrase and topoisomerase IV leading to less efficient binding of quinolone antibiotics [45]. This mechanism has been answerable for widespread quinolone resistance among the family Enterobacteriaceae.
Reduced Permeableness
The cytoplasmic membrane may be a barrier to water loving compounds. Entry of cytoplasmatically targeted compounds is typically through carrier-mediated transport mechanisms or via channels within the outer membrane of Gram-negative microorganism formed by porins (e.g. OprD porin) [46]. Antibacterial drug transported in this way could be subject to resistance by loss of non-essential transporters, by lack of porins or by mutations that are ready to modify the structure of these channels and therefore decreasing the inflow [47]. Some microbes possess impervious cell membranes that forestall drug inflow as exemplified by P. aeruginosa. For example, Neisseria gonorrhoea, the causative organism of gonorrhea [one of the foremost prevailing sexually transmitted disease], will gain antibiotic resistance by acquiring mutation within the gene encoding the membrane supermolecule ‘porin’, therefore inhibiting the transport of the antibiotics penicillin and tetracycline into the cell and rendering the cells immune against the effect of medication. Moreover, several massive molecule antimicrobials are naturally inactive against certain groups of bacteria because they simply cannot pass into the bacterial cell [48].
Efflux Systems in Microorganism
One of the foremost common drug resistance mechanisms is active efflux of medication from the bacterial cells to forestall the intracellular accumulation of venomous compounds [49]. Such drug resistant bacteria harbor energy driven drug efflux pumps that squeeze out antimicrobial agents therefore reducing their intracellular concentrations to sub or non-inhibitory levels by pumping such molecules out of the cell in a method that does not involve the alteration or degradation of the medication. The presence of a mechanism that decreases the buildup of tetracycline in E. coli was known in 1978 by Levy and McMurry (Fernández et al., 2012). Two years later, it was disclosed that resistance to tetracycline was due increased efflux and not to reduced inflow, as initially suggested [50]. Since then, several classes of efflux pumps are characterized in each Gram-negative and Gram-positive pathogens. These systems could also be substrate-specific [for a selected antibiotic like tet determinants for tetracycline and mef genes for macrolides in pneumococci] or with broad substrate specificity, that are sometimes found in MDR bacteria [51].
This mechanism of resistance affects a wide range of antimicrobial categories as well as protein synthesis inhibitors, fluoroquinolones, β-lactams, carbapenems and polymyxins. The genes encoding efflux pumps may be situated in mobile genetic parts (MGEs) or within the chromosome. Significantly, chromosomally encoded pumps can explain the inherent resistance of some bacterial species to selected antibiotic [52]. There are two main types of active efflux pumps. The primary kind, known as primary active transport, uses the reaction of nucleotide [ATP] to actively pump drugs from cells, whereas the second kind, known as secondary active transport, uses ion gradient for active drug efflux from cells. The ATP driven transporters are called ABC [for ATP binding cassette] or P-glycoprotein transporters [53]. Each transport systems are employed by microorganism to resist the repressive effects of antimicrobial agents and are typically noted as efflux pumps [54]. In addition to single drug efflux pump systems [55], bacteria may also express efflux pumps that are capable of squeezing out multiple structurally totally different antimicrobial agents and are called multidrug efflux pumps [56,57] and this has contributed considerably to increasing multidrug resistance in microorganism.
Antibiotic Inactivation by Microorganism Enzymes
Among many mechanisms concerned in the development of antibiotic resistance, drug modification plays a big role in rendering several therapeutically helpful medications useless. Some microorganism manufactures modifying enzymes that reside inside or close to the cell surface, that selectively target and inactivate the drug. Enzymatic inactivation either by hydrolysis or by modification (group transfer and redox mechanisms) is a major mechanism of resistance to natural antibiotics in pathogenic microbes [47]. The resistant isolates in most cases inherit the antibiotic resistance genes on resistance (R) plasmids. These resistance determinants are most likely acquired by pathogenic bacteria from a pool of resistance genes in different bacteria genera, as well as antibiotic producing organisms. No enzymes that hydrolyze or modify man made antimicrobials have been found [47]. Moreover, antibiotic inactivation mechanisms share several similarities with well characterised enzymatic reactions and resistance proteins show homologies to well-known metabolic and signalling enzymes with no antibiotic resistance activity. Therefore, one will speculate that these are the initial sources of resistance [58]. Either hydrolysis or group transfer reactions, or alternatively oxidization or reduction reactions, can sign for the inactivation mechanism. For example, Beta-lactamase, an enzyme elaborated by several Gram-positive and a few Gram-negative microorganism, converts antibiotic into penicilloic acid, that is therapeutically inactive. The production of the enzyme is inducible in Gram-positive microorganism whereas it is constitutive in Gram-negative bacteria. This reaction in addition forms the premise of chemical assay of the antibiotic since penicilloic acid may be titrated with iodine. The enzyme also provides a therapeutic target in the management of infections caused by penicillin resistant bacteria (Figure 1).
Acquired Resistance (Horizontal Gene Transfer)
The principal mechanism for the spread of antibiotic resistance is by horizontal transfer of genetic material. Antibiotic resistance genes could also be transferred by completely different mechanisms. Acquisition of foreign deoxyribonucleic acid material through HGT is one amongst the foremost necessary drivers of microorganism evolution and it is often accountable for the development of antimicrobial resistance. Most antimicrobial agents employed in clinical practice are [or derive from] product naturally found within the surroundings (mostly soil). Microorganism sharing the environment with these molecules harbors intrinsic genetic determinants of resistance and there is strong proof suggesting that such “environmental resistome” is a verdant source for the acquisition of antibiotic resistance genes in clinically relevant bacteria [39].
Moreso, this genetic exchange has been involved in the dissemination of resistance to several often-used antibiotics. Classically, bacteria acquire external genetic material through three main ways, i) transformation (incorporation of naked DNA), ii) transduction (phage mediated) and iii) conjugation (bacterial “sex”). Transformation is possibly the simplest type of HGT, however only a few of clinically relevant bacterial species are able to “naturally” incorporate naked DNA to develop resistance. Emergence of resistance within the hospital surroundings typically involves conjugation, a proficient technique of DNA transfer that involves cell-to-cell contact and is probably going to occur at high rates in the gastrointestinal tract of humans under antibiotic treatment. As a general rule, conjugation uses Mobile Genetic Components (MGEs) as vehicles to share valuable genetic data, although direct transfer from chromosome to chromosome has been well characterized [59].
The foremost necessary MGEs are plasmids and transposons, each of them plays a vital role in the development and dissemination of antimicrobial resistance among clinically relevant organisms. Finally, one amongst the foremost efficient mechanisms for accumulating antimicrobial resistance genes is delineated by integrons, which are site-specific recombination systems capable of recruiting open reading frames within the mobile gene cassettes. Integrons provide competent and rather easy mechanism for the addition of new genes into bacterial chromosomes, alongside the mandatory machinery to confirm their expression; a healthy strategy of genetic interchange and one of the main drivers of bacteria evolution.
Integron
Integrons are genetic elements that contain a site-specific recombination system able to capture, express and exchange specific DNA components, known as gene cassettes [60]. They have two main components. The first is made up integron integrase gene (intI) and its promoter (PintI), an integration named attI [attachment site of the integron), a constitutive promoter (Pc) for the gene cassettes integrated at the attI site and the second component are gene cassettes which are genetic units that embody one ORF, typically without a promoter, and specific recombination site, attC [61]. Recruiting exogenous genes represents a speedy adaptation against antimicrobial compounds, and therefore the integron useful platform seems dead suited to capturing those specific genes that enqble bacteria pathogens to face challenges posed by multiple antibiotic treatment regimes. It contributes to genomic malleability and environmental adaptation of bacteria [62]. Integrons are often thought about as one of the most important genetic carriers of and vectors for dissemination of antibiotic resistance determinants in bacteria [63].
Integron is classified based on their genomic context as mobile integrons, whenever they are related to transposons, or chromosomal integrons, once placed within the bacteria chromsome [63,64]. Mobile integrons are principally found in Gram-negative bacteria, though a number of studies have reported their presence amongst Gram-positive species [61,63].
Classes of Integrons
Based on genetic relatedness of the integrase gene, mobile integrons are grouped into five distinct classes with those carrying intI1 outlined as ‘class 1’, intI2 as ‘class 2’, intI3 as ‘class 3’, etc. intI1, intI2 and intI3 were first known in association with mobile genetic components and intI4 and others with chromosomal integrons [62]. Class one integrons were delineate in 1989 [60] and are reported as the most frequent and widespread, particularly in clinical settings. This category principally carries antimicrobial resistance genes and it is extremely disseminated because of the shut association with transposons, typically embedded in conjugative plasmids [63]. Complete integron are not mobile component itself because it lacks functions for self-mobility. However, individual gene cassettes are thought-about mobile within integron [64]. Moreover, integrons are usually embedded in larger Mobile Genetic Element (MGEs) like transposons and plasmids, which mediate horizontal dissemination of the integrons [65]. Multiple Resistant Integron (RIs) classes 1-3 integrons are capable of acquiring same gene cassettes via similar recombination platform, that had been supported by the in vitro removal and integration which occurs by means of recombination sites from such integrons [66].
Most of the presently obtainable studies on integrons had been conducted on class 1 integron, with focus on Gram-negative bacteria. As a definite form of integron, class 4 integron was first of all known on the some chromosome Vibrio cholerae and found to be an integral part of many γ-proteobacterial genomes [67], which had also been considered to be a leading concern on both antimicrobial resistance and bacterial genome evolution. The remaining classes of integrons may contain antibiotic resistance gene cassettes, however their worldwide prevalence remains low [66]. With the detection of the superintegron and thousands of cassettes entrapped in integrons of environmental species, we now have the tendency to currently have an improved understanding of the vast resources embedded among this system. However, many queries of importance concerning this system are still without satisfactory answers, together with insights concerning specific recombination processes, cassettes genesis, and cassettes exchange dynamics among advanced microbial populations.
Types of Integrons
In relevance to acquisition of antimicrobial resistance, two major teams of integrons are described: “Chromosomal Integrons” (Cis) and “Mobile Integrons” (Mis). When they are found on the chromosome of many bacterial species; actually, in silico analysis showed that 17% of sequenced bacterial genomes exhibit such genetic arrangements [63]. MIs are not self-permutable components however are found on mobile genetic components like plasmids and transposons, which assist in their dissemination among microorganism. MIs contain a restricted variety of GCs that is a smaller amount than ten GCs [68]. The GCs regions of those MIs typically encrypt antibiotic resistance determinants. MIs are thus generally known as “resistant integrons” (RIs) or “Multidrug Resistance Integrons” (MRIs).
Integrons and Antibiotic Resistance
Mobile integron choice and dissemination in microorganism and antibiotic pressure has played a serious role. One hundred thirty GCs conferring resistance to antibiotics and sixty GCs of unknown functions are delineating in MIs [69]. Genes concerned in resistance to the majority antibiotic families are embedded in GCs, together with beta-lactams, aminoglycosides, trimethoprim, macrolides, Chloromycetin, fosfomycin, lincosamides, rifampicin and quinolones [70]. Studies have advised that MIs were a lot of rife in microorganism communities subjected to direct or indirect antibiotic pressure in clinical, agricultural, and environmental settings [71,72]. Recent in vitro studies have shown that antibiotics area unit able to induce integrase transcription, each in CIs and MIs, via the SOS response. The SOS response may be a world regulative network controlled by the transcriptional represser LexA and elicited by stress resulting in direct or indirect deoxyribonucleic acid injury, like injury ensuing from exposure to some wide used antibiotics like fluoroquinolones, beta-lactams, trimethoprim and aminoglycosides [73,74].
The activation of the SOS response in microorganism leads to integrase over expression that result in the raise of GCs recombination events. Clinical, veterinary and environmental surveys have shown that microorganism harboring MIs area unit typically related to Multi Drug Resistant (MDR) phenotypes [75]. Recently, several studies are conducted and have documented the involvement of integrons in acquisition of antibiotic resistance in microorganism [76,77]. Integrons are involved in an exceedinglyiding antibiotic resistance in microorganism in a big selection of environments like contaminated water, poultry, food, hospitals, etc.
Bacterial Plasmid
The acquisition of novel genetic material from alternative resistant organisms is accountable for the resistance in some microorganism. The transfer of genetic material between identical microorganism species or completely different species, apart from the transfer through the parent to its progeny, is termed horizontal gene transfer (HGT). Microorganism can exchange the genetic material through the processes of transformation, conjugation and transduction. These processes of genetic transmission are expedited by a Mobile Genetic Element such as plasmid. Plasmids might carry resistant genes and transmit these to other microorganism [particularly Gram-negative bacteria] through conjugation. Throughout conjugation, pilus is created between two bacterail cells, through which the genetic material or plasmid carrying resistant genes are transferred [78]. The donor cell in the presence of pheromone produces a proteinaceous structure on the cell surface known as aggregation substance (AS) that binds to the enterococcal binding substance (EBS) present on the surface of the recipient.
A sexual union channel is produced between donor and recipient that allow the transfer of the plasmid deoxyribonucleic acid. Haven acquired the plasmid; the recipient stops the assembly of pheromone and initiates the assembly of an encoded inhibitor peptide that serves to desensitize the bacterial cell to an occasional level of endogenous and exogenous s pheromone formed by the donor. The pheromone-responsive conjugative plasmid system has been most extensively studied for plasmids [79]. R-factor found in enteric bacteria, were the foremost examples of horizontally transferred antibiotic resistance [80]. R-factors are quite advanced plasmids carrying not solely antibiotic resistance genes, however, additionally transfer functions permitting a speedy transmission among a population of connected Gram-negative bacteria. The resistance genes on these plasmids are either carried on plasmid integrated transposons or are inserted in an integron [80].
Bacteria Cell to Cell Transfer of R-Plasmids
R-plasmids are not solely steadily maintained, however also usually transferred between bacterial cells at a really high potency, in several cases approaching 100 percent. The molecular mechanism of this cell to cell transfer has been studied principally in E. coli and its relatives and within the plasmids of incompatibility cluster FI (F or sex plasmid) [81]. In recent years, the genes transferred were found to be homologs of the virB genes of Agrobacterium tumefaciens, which transfer a small piece of bacterial DNA into a plant nucleus, and a number of other effector protein injection systems of human and animal pathogens [82]. A sequence within the circular, double-stranded deoxyribonucleic acid of plasmids is recognized, and a single-stranded cut is formed by the VirD2/TraI enzyme with the enzyme protein covalently attached to the 5′-end of the cut strand. This complex is sometimes known as a relaxosome, because the nicking of one strand leads to the relaxation of supercoiling of the plasmid DNA.
The protein (single stranded) DNA complex is then exported by the “mating-pair formation” complex, composed of a dozen completely different proteins. Though one of the original strands of the R-plasmid DNA is transferred utterly into the recipient cells, within the donor cell this strand is replaced by a rollingcircle replication mechanism, so that the entire R-plasmids currently exist each in donor and recipient cells. Additionally, some R-plasmids are equipped with functions that antagonize the attack by restriction endonucleases [83]. Finally, it ought to be stressed that conjugational transfer of plasmids is not a completely unique mechanism invented for multidrug resistance; so, horizontal transfer of genes, to great extent through the conjugational mechanism, has been the most mechanism used for the evolution of the many teams of microorganism [84]. Gram-positive bacteria additionally perform cell to cell transfer of plasmids [85].
Transposons
Transposable elements (TEs) are deoxyribonucleic acid sequences that are able to alter their positions a genome, thereby producing new mutations and could alter the cell’s genome size and usually lead to duplication of the individual transponsable element [86]. In general, they are as junk DNA and they make up a large section of the C-value of the eukaryotic cell and are helpful in research to modify the DNA within the living organisms [87]. TEs possess two main characteristics that differentiate them from alternative genetic components. As they are mobile, they can vary their genetic surroundings and therefore changing the genetic environment of the locus into which they insert [88]. Since they need the intrinsic ability to multiply throughout the transposition scheme, they are routinely amplified, with a large copy number, restricted solely by the carrying capability of the genome [89]. Many sorts of transposable elements are present. One of which is the Mobile Genetic Elements which include two classes based on their mechanism of transposition and may best be delineate as both “copy and paste” (for class I TEs) or “cut and paste” (for class II TEs) [90].
Transposon class I [Retro-Transposons]
These transposons function by copying themselves from DNA to RNA by transcription and vice versa by reverse transcription. The ensuing DNA copy can after being inserted into the genome at a different position. The reverse transcriptase enzyme will initiate the reverse transcription, which is commonly coded for by the TE itself. Retro-transposons behave terribly equally to retroviruses, like HIV [90].
Transposon class II [DNA Transposons]
In this case, RNA intermediate is not required for the cut and paste transposition mechanism of class II TEs which are catalyzed by numerous sorts of transposase enzymes [88]. The transposase produces a staggered cut at the target site, forming sticky ends. The transposase then gets rid of the DNA and ligates it into the target site [90]. The DNA polymerase fills in the resultant gaps from the sticky ends and DNA ligase closes the sugar phosphate backbone. The ultimate outcome is target site duplication and production of short direct repeats at the insertion sites of DNA transposons. Therefore, gene duplications may result and is a crucial evolutionary process [70]. A variety of genetic elements are powerfully related to the exchange and transfer of antimicrobial resistance genes among Gram-negative microorganism. Transposons can transfer resistance genes intracellularly. Transposons like Tn3, Tn5, Tn7, Tn9 and Tn10, confers resistance to antibiotics, namely, Tn3 to ampicillin, Tn5 to kanamycin, Tn7 to trimethoprim, streptothricin, spectinomycin, streptomycin, Tn9 to chloramphenicol and Tn10 to tetracycline respectively . Therefore, wide array of transposons are involved in imparting resistance. Labar et al., [90], found the regional dissemination of a trimethoprim resistance gene cassette via a successful transposable element. They observed a 50% increase in the occurrence of trimethoprim resistance among fecal E. coli from healthy Nigerian students between 1998 and 2005. In this study 16.3% isolates carried transposons conferring trimethoprim resistance, indicating transposition as a principal mechanism for disseminating dihydrofolate reductase [dfrA7] among E. coli from Nigeria and Ghana.
How to Control or Reduce Antibiotic Resistance Development
The most serious outcome of antibiotics utilization is the development of bacteria resistant strains; this has aggravated nonstop efforts to exert management over antibiotic usage. It is clear that antibiotic resistance looks inevitable. However, there are steps which may be taken to forestall or at least delay this process. Years before now, many alternative solutions are imply by knowledgeable consultants and all major international health teams (e.g., WHO and CDC). Among the solutions for action are strict controls on antibiotic use by humans, requiring correct prescriptions [no use of antibiotics to treat colds and viral infections], no delivery of antibiotics without a doctor’s prescription (reducing unnecessary use of antibiotics) and controlled therapeutic use in animal husbandry and agriculture. Amusingly, the Swann recommendations of 1969 were the primary to incorporate a ban on nontherapeutic use in animals and agriculture, an affordable however extremely contentious suggestion that has been not possible to enforce in several countries to the current day.
Deception has played a role in this failure; several of the antimicrobials approved for treatment of humans are given to animals under the cover of various names for various uses. It is clear that restriction of antibiotic use is tough to implement on a world scale. Universal adherence to the suggested rules for restraint may have a positive result and however will surely not eliminate resistance. A lot of research geared towards knowing the molecular basis of resistance ought to be inspired.
The Danger Ahead
In the past few decades, because of the emergence of resistant strains, therapeutic advantage of a number of the potent antibiotics was utterly lost. Nearly twenty years, from the early 1980s to the late 1990s, not a single truly new antibiotic was introduced into clinical use. Only five new antibiotics were approved from 2003 to 2007, compared to the sixteen approved from 1983 to 1987. Treatment of Gram-negative infections is even dimmer because of the tight nature of the Gram-negative envelope and presence of multiple efflux pumps together with other resistance mechanisms contribute to the problem of this task. If the emergence of antibiotic resistance continues to follow this trend, it is not out of place to assume that resistance to novel antibiotics may emerge sooner than expected. Despite cautious usage of some antibiotics, the life span of a new antibiotic is cut short by the emerging resistance. It is evident that continues offer of structurally novel antimicrobial agents with multiple mode of action is required to combat the dilemma of drug resistance. The optimism related to antimicrobial peptides due to their broad-spectrum activity, absence of cross-resistance with the present antibiotics and low chance of developing resistance is hindered by their poor bioavailability and high cost involved in their manufacture. Physicians are typically taking resort to multiple antibiotic therapies, which are probably more effective than employing a single antibiotic. Scientists are searching for various ways e.g., enhancing the susceptibility of bacteria to antibiotics by using plant-derived antimicrobials [viz., thymol, carvacrol, cinnamaldehyde, allyl isothiocyanate]. Let us hope for an improved future while being aware of the unstable situation that is prevailing at present.
Conclusion
The importance and value of antibiotics cannot be overrated; we are totally dependent on them for the treatment of infectious diseases, and they ought not to be considered as mere commodities. In addition to their use in the treatment of infectious diseases, antibiotics are critical to the success of advanced surgical procedures, including organ and prosthetic transplants. In spite of all the good intentions to control antibiotic usage [but limited action], there is doubt that the situation with respect to antibiotic resistance can be solved. Resistance mechanisms are pandemic and has created enormous clinical and financial burden on health care systems worldwide. There are no simple solutions to the problem. Decisive actions that require significant commitment and enforcement are never popular, even if lives can be saved. Fortunately, not all bacterial pathogens are resistant all at the same time and many respond to empirical treatment with antimicrobial agents administered in the community. Success is perhaps due to luck rather than to good judgment. Multiple drug resistance is an unavoidable natural phenomenon, posing a serious worldwide menace to public health. Pathogens tend to adopt various resistance mechanisms to survive the unfavorable conditions. A cooperative action at global level is a must to combat the microbial multidrug resistance.
Recommendations
Improved knowledge of molecular mechanisms controlling MDR ought to facilitate the development of novel therapies to combat these stubborn infections and will help cultivate a deeper understanding of the pathobiology of microorganism. A radical understanding of the molecular mechanisms involved in the emergence and dissemination of MDR, careful experimental validation and enhanced surveillance systems are keys for safety and effectiveness of antibiotics against multidrug resistance. Additional studies of the molecular basis of Gram-negative bacteria resistance in our region are required to achieve an improved understanding of the complexity of resistance mechanisms in these organisms. Given the various considerations, the best one can expect is that all physicians and health care centers provide their patients with environments that are resistance free by taking stricter measures in infection control and antibiotic use. This must be backed up by efforts to prevent dumping of antibiotics into the environment through sewer systems; complete destruction of antibiotics before disposal should be common practice. The finding that 38% of isolates revealed no resistance mechanism to explain the carbapenem resistance highlights the urgent demand for more studies to investigate the unknown resistance mechanisms conferring carbapenem resistance to Gram-negative bacteria.
References
- Maranan MC, Moreira B, Boyle Vavra S (1997) Antimicrobial resistance in Staphylococci: Epidemiology, molecular mechanisms, and clinical relevance. Infect Dis Clin North Am 11 :813-849.
- Zaman S, Hussain M, Nye R (2017) A Review on Antibiotic Resistance: Alarm bells are ringing. Cureus 9(6): 1-8.
- Aminov RI (2010) A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1: 134.
- Davies J, Davies D (2010) Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417-433.
- Normark BH, Normark S (2002) Evolution and spread of antibiotic resistance. J Intern Med 252: 91-106.
- Iwu MW, Duncan AR, Okunji CO (1999) New antimicrobials of plant origin. In Perspectives on New Crops and New Uses: Janick, J, Ed. ASHS Press: Alexandria, VA, USA, pp. 457-462.
- Bush K (2004) Antibacterial drug discovery in the 21st Clin Microbiol Inf 10: 10-17.
- (2002) World Health Organization (WHO) Antimicrobial Resistance: Fact Sheet No. 194: WHO: Geneva, Switzerland.
- Woodford N, Ellington MJ (2007) The emergence of antibiotic resistance by mutation. Clin Microbiol Infect 13: 5-18.
- Bennett PM (2008) Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British Journal of Pharmacology 153(1): 347-357.
- Beović B (2006) The issue of antimicrobial resistance in human medicine. International Journal of Food Microbiology 112(3): 280-287.
- Aminov RI (2011) Horizontal gene exchange in environmental microbiota. Frontier Microbiology 26(2): 158.
- Singh V (2013) Antimicrobial resistance in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education. Formatex Research Center 1: 291-296.
- Popęda M, Płuciennik E, Bednarek AK (2014) Proteins in cancer resistance. Postępy Higieny i Medycyny Doświadczalnej 68: 616- 632.
- Engidaw A, Betelihem T, Solomon T (2016) A review on molecular mechanisms of bacterial resistance to antibiotics. European Journal of Applied Sciences 8 (5): 301-310.
- Aharon A, Venezia S, Muntz O, Krichali T, Igra Y, et al. (2005) Multidrug resistant Acinetobacter baumannii. Emerg Infect Dis 11(1): 22-29.
- Federico P, Hujer A, Hujer K, Decker B, Rather P, et al. (2007) Global challenge of multidrug resistant Acinetobacter baumannii. Antimicrobial agents and chemotherapy pp. 3471-3484.
- Aires de Sousa M, MI Crisóstomo, I Santos Sanches, JS Wu, J Fuzhong, et al. (2003) Frequent recovery of a single clonal type of multidrug-resistantStaphylococcus aureus from patients in two hospitals in Taiwan and China. J Clin Microbiol 41(1): 159-163.
- Richard H (2007) Emerging options for treatment of invasive, multidrug-resistant Staphylococcus aureus infections. The Journal of Human Pharmacology and Drug Therapy 27: 227-249.
- Boyce JM, Opal SM, Chow JW, Zervos MJ, Potter Bynoe G, et al. (1994) Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistant. Journal of Clinical Microbiology, 32: 1148-1153.
- Murray BE (1998) Diversity among multidrug-resistant enterococci. Emerging infectious diseases, 4(1): 37-47.
- Bostic GD, Perri MB, Thal LA, Zervos MJ (1998) Comparative in vitro and bacterial activity of oxazolidinone antibiotics against multidrug resistant enterococci. Diagnostic Microbiology and Infectious Disease 30: 109-112.
- Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton NR, et.al (2006) The multidrug resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nature genetics 38: 779-786.
- Jing Jou Y, Ko WC, Tsai SH, Wu HM, Wu JJ (2001) Outbreak of Infection with Multidrug resistant Klebsiella pneumoniae carrying blaIMP-8 in a University Medical Center in Taiwan. J Clin Microbiol 39: 4433-4439.
- Elizabeth BH, Tam VH (2010) Detection and treatment options for Klebsiella pneumonia carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother 65 (6): 1119-1125.
- Whitney CG, Farley MM, Hadler J, Harrison LH, Lexau C, et al. (2000) Increasing Prevalence of multidrug resistant Streptococcus pneumoniae in the United States. N Engl J Med 343: 1917-1924.
- Poudel, A, Chie, N, Yukari, F, Haruka, S, Basu, et al. (2012) Molecular characterization of multidrug resistant Mycobacterium tuberculosis isolated in Nepal. Antimicrobial Agents and Chemotherapy 56(6): 2831-2836.
- Janak K (2011) Multidrug resistant Salmonella enterica. The Lancet Infectious Diseases 11 (11): 808 - 809.
- Jyoti T, Shrayanee D, Zeeshan F, Saif H (2014) Multidrug resistance: An emerging crisis. Interdisciplinary Perspectives on Infectious Diseases p. 7.
- Fishbain, Peleg AY (2010) Treatment of Acinetobacter infections. Clinical Infectious Diseases 51(1): 79-84.
- Kahne D, Leimkuhler C, Lu W, Walsh C (2005) Chemical Reviews 105: 425-448.
- Loeffler J, Stevens, DA (2003) Antifungal drug resistance. Clinical Infectious Diseases 36(1): 31-S41.
- Walsh C, (2000) Molecular mechanisms that confer antibacterial drug resistance. Nature 406: 775-781.
- Wright GD (2005) Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv Drug Deliv Rev 57: 1451-1470.
- Giguère S (2006) Antimicrobial Drug Action and Interaction: An Introduction. Antimicrobial therapy in Veterinary Medicine (4th). Ames Iowa, USA.
- Koronakis V, Eswaran J, Hughes C (2004) Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu Rev Biochem 73: 467-489.
- Fernandez L, Hancock RE (2012) Adaptive and mutational resistance: role of porins and efflux pump in drug resistance. Clin Microbiol Rev 25: 661-681.
- Forbes BA, Sahm, DF, Weissfeld AS (1998) Bailey and Scott’s Diagnostic Microbiology, (10th Edn). Mosby Inc, St. Louis Missouri, USA G.D.
- Jose MM, Cesar AA (2016) Mechanisms of Antibiotic Resistance. Microbiol Spectr 4(2): 1-37.
- Lambert PA (2005) Bacterial resistance to antibiotics: modified target sites. Adv Drug Deliv Rev 57: 1471-1485.
- Fabrega A, Madurga S, Giralt E, Vila J (2009) Mechanism of action of and resistance to quinolones. Microbial biotechnology 2: 40-61.
- Drapeau CM, Grilli E, Petrosillo N (2010) Rifampicin combined regimens for gram-negative infections: data from the literature. International journal of antimicrobial agents 35: 39-44.
- Widdowson CA, Klugman KP (2008) The molecular mechanisms of tetracycline resistance in the pneumococcus. Microbial drug resistance 4: 79-84.
- Robicsek A, Jacoby GA, Hooper DC (2006) The worldwide emergence of plasmid-mediated quinolone resistance. The Lancet infectious diseases 6: 629-640.
- Silver LL (2003) Novel inhibitors of bacterial cell wall synthesis. Curr Opin Microbiol 6: 431-438.
- Yoneyama H, Katsumata R (2006) Antibiotic resistance in bacteria and its future for novel antibiotic development, Biosci Biotechnol Biochem 70: 1060-1075.
- Rachakonda S, Cartee L, (2004) Challenges in antimicrobial drug discovery and the potential of nucleoside antibiotics, Curr Med Chem 11: 775-793.
- Webber MA, Piddock LJ (2003) The importance of efflux pumps in bacterial antibiotic resistance. The Journal of antimicrobial chemotherapy 51: 9-11.
- Fernández L, Robert EWH (2012) Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clinical Microbiology Reviews 25(4): 661-681.
- Poole K (2005) Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 56(1): 20-51.
- Singh V, Weinstock GM, Murray BE (2002) An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 46: 1845-1850.
- Davidson AL, Maloney PC (2007) ABC transporters: how small machines do a big job. Trends in microbiology 15: 448-55.
- Levy SB (2002) Active efflux, a common mechanism for biocide and antibiotic resistance. Journal of applied microbiology 92: 65S-71S.
- Alekshun MN, Levy SB (2007) Molecular mechanisms of antibacterial multidrug resistance. Cell 128: 1037-1050.
- Kerr ID, Jones PM, George AM (2010) Multidrug efflux pumps: the structures of prokaryotic ATP-binding cassette transporter efflux pumps and implications for our understanding of eukaryotic P-glycoproteins and homologues. The FEBS journal 277: 550-563.
- Bhardwaj AK, Mohanty P (2012) Bacterial efflux pumps involved in multidrug resistance and their inhibitors: rejuvinating the antimicrobial chemotherapy. Recent patents on anti-infective drug discovery 7: 73-89.
- Manson JM, Hancock LE, Gilmore MS (2010) Mechanism of chromosomal transfer of Enterococcus faecalis Pathogenicity Island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci 107(27): 12269-12274.
- Domingues S, Gabriela JS, Nielsen KM (2015) Global dissemination patterns of common gene cassette arrays in class 1 integrons. Microbiology 161: 1313-1337.
- Domingues, S, da Silva, G. J, Nielsen, K. M (2012a) Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mobile Genet Elements 2: 211-223.
- Boucher, Y, Labbate, M, Koenig, J. E, Stokes, H. W (2007) Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol, 15, 301-309.
- Cambray G, Guerout AM, Mazel D (2010) Integrons. Annu Rev Genet 44: 141-166.
- Mazel D (2006) Integrons: agents of bacterial evolution. Nat Rev Microbiol, 4: 608-620.
- Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti Bragadin C, et al. (2002) Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob Agents Chemother 46: 3665-3668.
- Yang D, Xuerui B, Lili J, Lei C, Junyan L, et al. (2015) Resistance integrons: class 1, 2 and 3 Integrons. Ann Clin Microbiol Antimicrob 14(45): 1-11.
- Martín BS, Lapierre L, Cornejo J, Bucarey S (2008) Characterization of antibiotic resistance genes linked to class 1 and 2 integrons in strains of Salmonella spp. isolated from swine. Can J Microbiol 54: 569-576.
- Naas T, Mikami Y, Imai T, Poirel L, Nordmann P (2001) Characterization of In53, a class 1 plasmid and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. Journal of Bacteriology 183: 235-249.
- Partridge SR, Tsafnat G, Coiera E, Iredell JR (2009) Gene cassettes and cassette arrays in mobile re- sistance integrons. FEMS Microbiology Reviews 33: 757-784.
- Iyer A, Elie, B Esam, A Alaaeddin, AES Hani, M AH, et al. (2013) Transposable elements in Escherichia coli antimicrobial resistance. Advances in Bioscience and Biotechnology 4: 415-423.
- Barlow RS, Fegan N, Gobius KS (2009) Integron containing bacteria in faeces of cattle from different pro- duction systems at slaughter. Journal of Applied Microbiology 107: 540-545.
- Kristiansson E, Fick J, Janzon A, Grabic R, Rut gersson C, et al. (2011) Pyrosequencing of antibiotic contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6(2).
- Baharoglu Z, Bikard D, Mazel D (2010) Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLOS Genetics 6.
- Cambray G, Sanchez Alberola N, Campoy S, Guerin E, Da Re S, et al. (2011) Prevalence of SOS-me- diated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mobile DNA 2(1): 6.
- Laroche E, Pawlak B, Berthe T, Skurnik D, Petit F (2009) Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France) FEMS Microbiology Ecology 68: 118-130.
- Soufi L, Abbassi MS, Saenz Y, Vinue L, Somalo S, et al. (2009) Prevalence and diversity of integrons and associated resistance genes in Escherichia coli isolates from poultry meat in Tunisia. Foodborne Pathogens and Disease 6: 1067-1073.
- Koczura R, Mokracka J, Jablonska L, Gozdecka E, Kubek M, et al. (2012) Antimicrobial re- sistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and ri- ver water. Science of the Total Environment 414: 680- 685.
- Harish C, Parul B, Archana Y, Babita P, Abhay PM, et al. (2017) Antimicrobial Resistance and the Alternative resources with special emphasis on plant based antimicrobials: A review. PlantS 6(16): 1-11.
- Wardal E, Sadowy E, Hryniewicz W (2010) Complex nature of Enterococcal pheromone-responsive plasmids. Pol J Microbiol 59: 79-87.
- Schröder G, Lanka E (2005) The mating pair formation system of conjugative plasmids: A versatile secretion machinery for transfer of proteins and DNA. Plasmid 54: 1-25.
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol 59: 451-85.
- Less M, Lanka E (1994) Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77: 321-324.
- Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299-304.
- Grohmann E, Muth G, Espinosa M (2003) Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev 67: 277-301.
- Shapiro JA (2010) Mobile DNA and evolution in the 21st Mobile DNA 1(1): 4.
- Bao W, Jurka MG, Kapitonov VV, Jurka J (2009) New superfamilies of eukaryotic DNA transposons and their internal divisions. Molecular Biology and Evolution 26: 983-993.
- Wicker T, Sabot F, Hua Van A, Bennetzen JL, Capy P, et al. (2007) A unified classification system for eukaryotic transposable elements. Nature Reviews Genetics 8: 973-982.
- Hua Van A, Rouzic AL, Boutin TS, Filee J, Capy P (2011) The struggle for life of the genome’s selfish architects. Biology Direct 6: 19.
- Kapitonov VV, Jurka J (2008) A universal classification of eukaryotic transposable elements implemented in Repbase. Nature Reviews Genetics 9: 411-412.
- Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, et al. (2008) Revised nomenclature for transposable genetic elements. Plasmid 60: 167-173.
- Reznikoff WS (2008) Transposon Tn5. Annual Review of Genetics 42: 269-286.
- Labar AS, Millman JS, Ruebush E, Opintan JA, Bishar RA, et al. (2012) Regional dissemination of a trimethoprim-resistance gene cassette via a successful trans- posable element. PLoS One 7.

 Review Article
Review Article