Abstract
Neurological abnormalities are included in the main effects of chronic lower level arsenic exposure in humans due to the consumption of contaminated drinking water. Recently, epigenetic mechanisms may explain the effects of xenobiotics. Arsenic exposure alters DNA methyltransferase (DNMT) enzyme expression and activity. It was interesting to determine if arsenic can modify other epigenetic mediators. Recently, the microRNA has been shown to regulatory effects at several molecular targets and regulate physiological processes. We performed a study to determine whether hVM1 cells exposed to arsenic during differentiation have altered expression levels of miR-125b,-148a and -132, and if these changes are associated with gene expression changes in nestin, CREB, BNDF and DNMT3b using real time RT-PCR. Statistically significant changes were observed in miR-125b and miR-148a expression. Although hVM1 cells exposed to arsenic showed a trend to downregulate miR-132, this was not significant. Nestin, an important marker of diminished differentiation, was negatively correlated with miR-125b and showed a trend for increased expression that may have a physiological interpretation. In conclusion, these results show that arsenic exposure could lead changes in the expression of key microRNA and therefore could affect neural differentiation in hVM1 cells.
Keywords: Epigenetics; Neural Differentiation; MiRs; Arsenic; HVM1 cells
Introduction
Chronic arsenic exposure is a global environmental health problem that affects millions of people worldwide. There are several regions in which drinking water exceeds the Environmental Protection Agency (EPA) limit of 10 ppb (10μg/L) and the local limit of arsenic (NOM-127 25μg/L). Several long-term health effects of low dose arsenic exposure in humans have been reported; the most important of these are the association with an increased risk of epithelial cancers of the skin, bladder, lung, liver, kidney and prostate, cardiovascular diseases, genotoxic and immunotoxic effects and, interestingly, neurological abnormalities [1-4]. Furthermore, arsenic exposure through drinking water induces general embryotoxicity and damages embryonic neurogenesis [5- 7] During embryogenesis, arsenite exposure might negatively affect the proliferation, survival and differentiation of embryonic neural stem cells [8]. The mechanisms that have been attributed to arsenic involve a wide range of effects; although debate continues, there is evidence to support the disruption of signaling cascades [9,10], oxidative stress [11] and epigenetic changes [12,13]. Recently, the field of epigenetics has opened up, allowing researchers to examine molecular mechanisms in response to gene-environment interactions and to elucidate how toxic compounds have broad effects. The wide effects associated to arsenic exposure suggest that arsenic is able to modify the responses through epigenetic mechanism. Epigenetic mechanisms allow cells to rapidly alter their long-term transcriptional activity, thereby permitting coordinated changes in gene expression without permanently altering the sequence of DNA. DNA methylation is one of the most frequent epigenetic alterations described following exposure to arsenic. It has been reported that arsenic is able to alter the activity of DNA methyltransferase (DNMT). DNMTs catalyze methyl group transfer from S-adenosylmethionine (SAM) into the C5´position of cytosine at CpG dinucleotides to produce 5-methylcytosine on promoter regions of genes; this silences gene expression [14,15]. The inhibition of DNMT activity by arsenic exposure appears to be a mechanism related to whole-genome and gene-specific DNA demethylation. However, in contrast, more recent reports have shown that localized DNA hypermethylation due to arsenic exposure is challenging to explain. [16] showed that localized DNA hypomethylation and hypermethylation occur following chronic arsenic exposure in Wistar rats. They observed a significant increase in the non-methylated form of the PP1 gene promoter, both in the cortex and hippocampus. At the same time, they observed an increase in the methylated form of the RELN gene promoter. Therefore, arsenic exposure may alter the methylation pattern of neural plasticity genes, but the mechanisms involved may be additional to or complemented by DNMT activity and methyl group consumption.
On the other hand, epigenetics has been recently been extended to microRNA (miR), which are small non-coding RNA molecules of about 21-23 nucleotides that function is posttranscriptional negative regulation of gene expression [17]. They have been implicated in the control of every cellular physiological process, including neural differentiation and maintenance of the neural phenotype [18,19] as well as in brain development and function [20-22]. miR-125b is highly expressed in the central nervous system, and is upregulated during neural differentiation in embryonic stem cells [23,25] demonstrated that miR-125b suppresses the expression of nestin (a marker of multipotent neural stem cells) by binding to the 3´-UTR of its mRNA, such that the expression of this target is downregulated when miR-125b is upregulated. Another miR investigated in the present study is miR- 132, which plays role in neural development and neural activity and is able to negatively modulate neural differentiation by regulating various genes [25,26]. In addition, miR-148a is an important regulator of DNMT3b expression [27], which is involved in the establishment of new methylation patterns during development and is most important during neural differentiation [28]. Evidence that chemical carcinogen exposure is capable of altering miRNA expression continues to emerge [29,30]. However, if miR-125b, miR- 132 and 148a expression can be altered by arsenic exposure, the implications for neural differentiation remain unknown. A new tool to evaluate the effect of arsenic exposure on neural differentiation is a stable cell line of human neural stem cells derived from the ventral mesencephalon (hVM1). hVM1 cells were isolated from a 10-weekold aborted human fetus, and immortalization was carried out by infection with a retroviral vector coding for v-myc (LTR-vmyc- SV40-Neo-LTR). This polyclonal cell line has the capacity for selfrenewal and is able to differentiate into neurons, astrocytes and oligodendrocytes [31-33]. In this study, we investigated the effect of arsenic exposure on miR-125b, miR-132 and miR-148a expression and additionally some target genes related to neural differentiation (nestin, CREB, BNDF and DNMT3b) in immortalized hVM1 cells.
Experimental Procedures
Cell Culture
hVM1 cells were cultured on 10μg/mL polylysine (PLL)- pretreated plastic ware in proliferation media (Dulbecco’s modified Eagle’s medium/F12 medium with epidermal growth factor and basic fibroblast growth factor (20ng/mL each; R&D System), Glutamax (Invitrogen), 1% Albumax (Invitrogen), 50mM HEPES (Invitrogen), 0.6% glucose, N2 supplement (Invitrogen), 1X nonessential amino acids, and penicillin/streptomycin). To induce cell differentiation, cells were seeded at 105 cells/cm2 in proliferation medium, and after 24 hours, the proliferation medium was replaced by differentiation medium (without epidermal growth factor and basic fibroblast growth factor and containing 1 mM dibutyryl-cAMP (Sigma) and 2ng/ml human recombinant glial cellderived neurotrophic factor (Preprotech)). Differentiation medium was changed every second day until the end of the experiment. Cells proliferated and differentiated at 37°C and 95% humidity in a low oxygen atmosphere (5% O2, 5% CO2, in a dual CO2/O2 incubator (Thermo Scientific)).
Cell Viability Assay
The resazurin reduction method was used for the determination of cell viability. The method is based on the reduction of resazurin, a normally non-fluorescent compound, to resofurin, a fluorescent metabolite, due to the highly reducing milieu of a living cell [34]. The assay was performed in 96-well plates. First, the number of cells to seed and the incubation time with resazurin was determined for hVM1 cells. Briefly, 3 X 104 cells were seeded onto 96-well plates in proliferation media, and after 48 hours the proliferation medium was replaced by differentiation medium in order to allow the attachment of cells; then, the vehicle, positive control and toxicant diluted in differentiation media was added at 5, 10, 50, 100 and 1000μg/L of sodium arsenite for 24 hours. After exposure, at day 7 of differentiation, cells were incubated for 2 hours in differentiation media containing 30% resazurin (30μg/ mL). The fluorescence of each condition (directly proportional to the number of metabolically active cells) was measured at 560nm (excitation)/590 nm (emission) using a Flex Station II TM fluorometer (Molecular Devices, Sunnyvale, CA) and the results of three independent experiments with triplicates were analyzed and plotted. The concentration of 5μg/L had cell viability above 80% and this used for subsequent experiments.
Determination of The Expression of Micrornas By Real- Time PCR
microRNA expression was determined for three separate experiments from duplicate cultures of hVM1 cells at 7 days of differentiation after exposure to 1 and 5μg/L sodium arsenite. microRNA were isolated using the mirVana Isolation Kit (Ambion, Austin, Texas), quantified and transcribed into cDNA with the specific TaqMan@MicroRNA Reverse Transcription Kit and TaqMan@MicroRNA Assay Kit (Applied Biosystems, Foster City, California, E.U.A) for miR-125b, miR-132, miR-148 were used for the amplification which performed on a CFX96 Touch thermocycler (Bio-Rad). The relative gene expression was obtained through 2-ΔΔCt method and U6 gene expression was housekeeping.
Determination of Target Gene Expression by Real-Time PCR
Cultures of hVM1 cells at 7 days of differentiation after exposure to 1 and 5μg/L sodium arsenite were collected and washed with PBS for RNA isolation using the Trizol method. The concentration and quality were determined by spectrophotometric analysis using a Synergy HT Multi-Mode Microplate Reader, and Gen5™ data analysis software (BioTek Instruments, Inc.). The samples were stored at -80°C until use. Complementary cDNA was synthesized with 1μg of total RNA using reverse transcriptase superscript II and reverse transcription reagents (Invitrogen) under the following conditions: 25°C 10 min, 35°C 90 min, 94ºC 5 min and 4°C 5 min. The qPCR mix included 1μl of cDNA (100 ng/μl), 0.1μl sense and antisense primers (20 pM) (Invitrogen) and 1X SYBR® Green PCR Master Mix (Applied Biosystems by Life Technologies) in a total volume of 10μl. Primers were designed and checked for specificity by a BLAST search. The primers used were: 5´- GGACTCGTTCAGAAAGCCCA-3 (fwd) and 5´- CTCTGGTTGCGTGTTGTTGG-3´ (rev) for DNMT3b, 5´- TGGAGCAGGAGAAACAGG-3’ (fwd) and 5’- CAGGTGTCTCAAGGGTAGC-3´ (rev) for nestin, 5´- ACTCTGGAGAGCGTGAA TGG -3´ (fwd) and 5´- ATCCAACAGCTCTTCTATCACG-3´(rev) for BDNF, 5´- GATCCGGGCCGTGAACGAAA-3´ (fwd) and 5´-TCTGAACTGTTTGGACTTGTGGAG-3´ (rev) for CREB and 5’-CGGCTACCACATCCAAGGAA-3´(fwd) and 5´- CGGCTACCACATCCAAGGAA-3´ (rev) for 18S that was used as housekeeping gene. The expression of target genes was determined using a CFX96 BioRad thermocycler (Bio-Rad). Cycling conditions were determined experimentally for each primer pair. The melting curve was analyzed with CFX Manager Software. DNMT3b, CREB, nestin and BDNF expression was normalized to 18S expression and data are expressed as the fold change using the 2−ΔΔCt method.
Statistical Analysis
For each set of data, it was determined if there was a normal distribution using the Shapiro-Wilk statistical test. For analysis of the viability assay, non-parametric Kruskal-Wallis and Dunn’s post-hoc test were performed. To determine the expression of miRs and genes, non-parametric one-way Kruskal-Wallis and Dunn’s post-hoc tests were performed. All analysis was performed using STATISTICA software 10.0 and the results were considered significant at p<0.05.
Results
Viability Assay
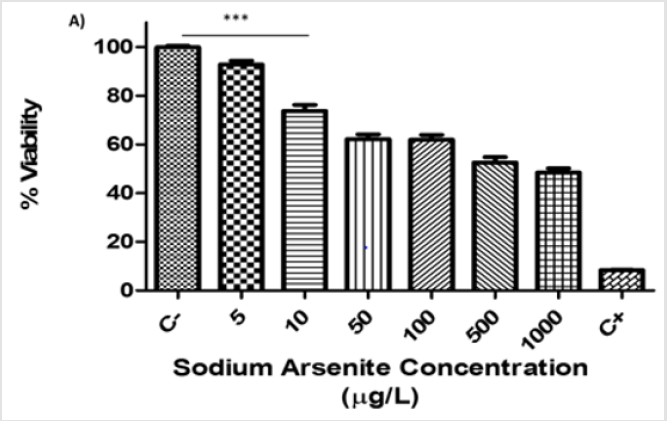
The viability assay was performed in hVM1 cells at 7 days of differentiation using the resazurin method. The hVM1 cells were treated with different concentrations of sodium arsenite (5-1000μg/L) for 24 hours. We observed that only at an arsenic concentration of 5μg/L were cells able to maintain viability above 80%; this was the concentration used for subsequent experiments. Above 10μg/L of sodium arsenic, the cell viability declined in a dose-dependent manner and was significantly lower than 80% (p<0.001) (Figure 1).
Figure 1: Viability assays of hVM1 cells exposed to sodium arsenite during differentiation (A). Percentage of cell viability of hVM1 cells in the presence of increasing doses of sodium arsenite (5-1000μg/L) at day 7 of differentiation. Data were analyzed by the means of the Kruskal-Wallis test and Dunn’s post-hoc test when significant values were obtained (*p<0.05 and ***p<0.001). Results of three independent experiments with triplicates, bars show mean+SEM.
(C-) Negative control, (C+) Positive control.
Expression of miR-132 and CREB in hVM1 cells Exposed To sodium arsenite
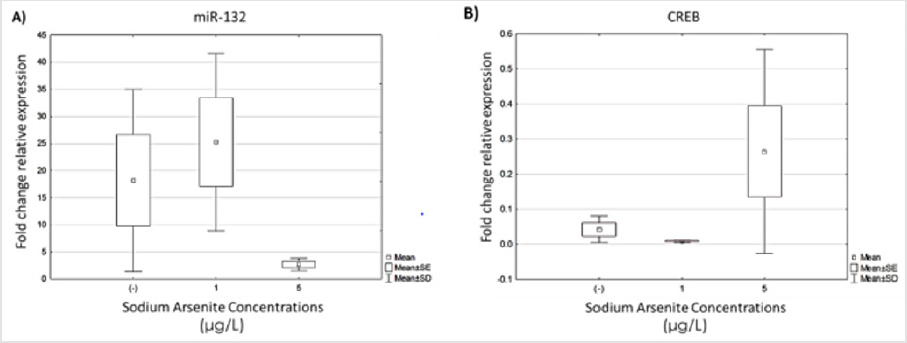
Once the sodium arsenite concentration at which hVM1 cells maintained greater than 80% viability was determined, we evaluated the expression of miR-132 and its transcription factor CREB in cultures exposed to 1 and 5μg/L of arsenic in both undifferentiated cultures and after 7 days of differentiation using qRT-PCR. Five independent experiments with duplicates of each condition were performed and the results are represented as the fold change between undifferentiated and differentiated cells. Although there were no significant differences between treated and untreated cells, we observed less dispersion in the data from hVM1 cultures exposed to 5μg/L of arsenic and a trend toward decreased expression of miR-132 in cultures after 7 days of differentiation compared to untreated cells (Figure 2A). We observed that the expression of the transcription factor CREB exhibited the opposite behavior to miR-132 expression (Figure 2B).
Figure 2: Analysis of miR-132 and CREB expression in hVM1 cells exposed to sodium arsenite. Relative expression levels of: A) miR-132 and B) CREB. Data were analyzed by the medians of the Kruskal-Wallis test and Dunn’s post-hoc test when significant values were obtained. Results of five independent experiments with duplicates. (-) Negative control.
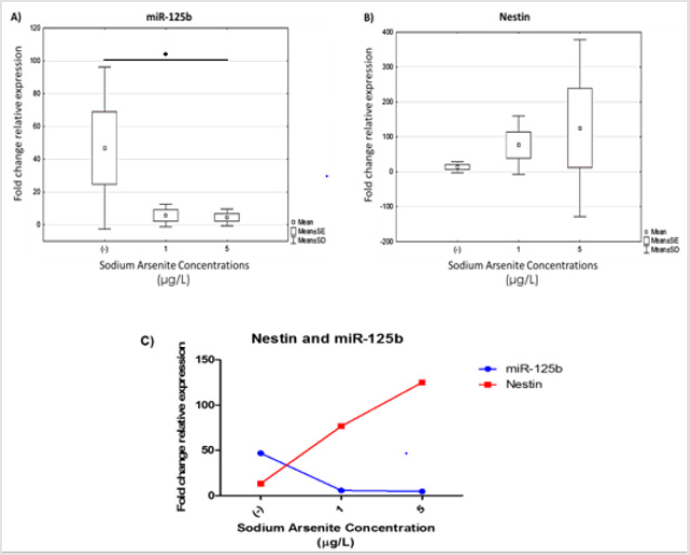
Decreased miR-125b Expression in hVM1 Cells Exposed to Sodium Arsenite during Differentiation is Related to Augmented Nestin Gene Expression
Figure 3: miR-125b expression and relationship with augmented nestin gene expression in hVM1 cells exposed to sodium arsenite during differentiation. Relative expression of: A) miR-125b and B) nestin. Data were analyzed by the medians of the Kruskal-Wallis test and Dunn’s post-hoc test when significant values were obtained (* p<0.05 and *** p<0.001). Results of five independent experiments with duplicates. C) Correlation of miR-125b and nestin expression in hVM1 cells treated with sodium arsenite during differentiation quantified by real-time PCR using specific TaqMan primers. Data represent the mean of five independent experiments with duplicates. (-) Negative control.
It has been demonstrated that dynamic changes in the expression of miRs occurs during neural differentiation, so to understand the effect of arsenic on miR-125b expression in the regulation of neural differentiation, the expression of miR-125b and a target gene (nestin) were assessed by real-time PCR. The results show that miR-125b was downregulated in both exposed cultures (1 and 5μg/L) and statistical differences were observed between untreated and arsenic exposed cultures (Figure 3A). At the same time, nestin gene expression was enhanced, which has been described like target gene of miR-125b (Figure 3B). We found that miR-125 and nestin gene expression showed opposing behavior, i.e. in untreated cultures, miR-125b expression was higher than in arsenic exposed cells. Moreover, and the expression of nestin was augmented in differentiation cultures following arsenic exposure, which could be a discreet effect of miR regulatory expression (Figure 3C).
BDNF Expression in Sodium Arsenite Exposed Hvm1 Cells During Differentiation
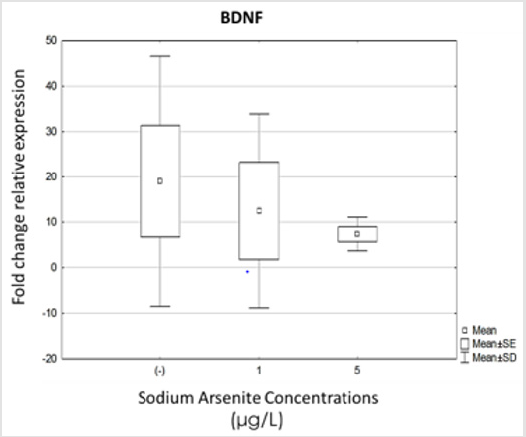
Because BDNF is one of the most important neuromodulators of neural differentiation and function, the analysis of its expression was included in this study. We evaluated BDNF expression under differentiation conditions and in cells exposed to 1 and 5μg/L of sodium arsenite. As we expected, BDNF expression was augmented in untreated differentiation cultures, but there were no statistically significant differences when hVM1 cells were exposed to sodium arsenite during differentiation (Figure 4).
Figure 4: Analysis of BDNF expression in hVM1 cells exposed to sodium arsenite. Relative expression levels of: A) BDNF. Data were analyzed by the medians of the Kruskal-Wallis test and Dunn’s post-hoc test when significant values were obtained. Results of five independent experiments with duplicates, bars show median with range. (-) Negative control.
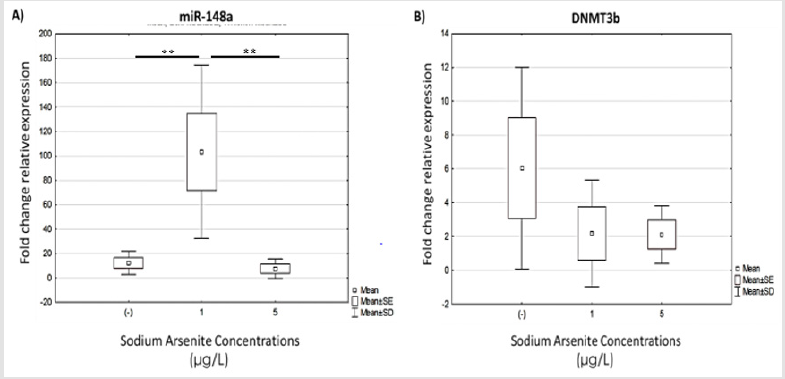
Hormesis Of Mir-148a Expression in Hvm1 Cells Following Sodium Arsenic Exposure
Figure 5: Analysis of BDNF expression in hVM1 cells exposed to sodium arsenite. Relative expression levels of: A) BDNF. Data were analyzed by the medians of the Kruskal-Wallis test and Dunn’s post-hoc test when significant values were obtained. Results of five independent experiments with duplicates, bars show median with range. (-) Negative control.
Finally, we analyzed miR-148 and DNMT3b (a target gene) expression in cells exposed to sodium arsenite during differentiation. miR-148a expression was significantly increase in cultures exposed to low doses (1μg/L) of arsenic compared to the other culture conditions, i.e. untreated and cells exposed to 5μg/L of arsenic (Figure 5A). We expected to find greatly diminished expression of DNMT3b in the 1μg/L arsenic cultures, but DNMT3b expression in the exposed groups was not significantly different compared to the control condition. However, we observed a trend for downregulation in both exposed cultures (1 and 5μg/L) (Figure 5B).
Discussion
During the development of the central nervous system, neural stem cells are able to generate neurons, astrocytes and oligodendrocytes, and therefore play important roles in the fetal and early postnatal stages. Growing evidence has been published regarding transplacental arsenite exposure; during embryogenesis, arsenic negatively affects the proliferation and differentiation of neural stem cells and leads to defects in the development and function of the central nervous system that could be permanent and associated with subsequent neural disorders [33,34]. However, in vitro studies of the effects of arsenic exposure on neural stem cells are scarce. In this study, we evaluated the effect of sodium arsenite on cell viability and the expression of three keys miR during in vitro differentiation in hVM1 cells. The viability assay showed that hVM1 cells are very sensitive to arsenic exposure, as sodium arsenite at 7 days of differentiation significantly decreased cell viability in a dose-dependent manner [34]. reported a Cytotoxicity Index (IC50) of 0.3mg/L inorganic arsenic at 72h in C17.2 cells (a neural progenitor cell line). They also observed viability percentages higher than ours at similar concentrations of 100, 500 and 1000μg/L inorganic arsenic. However, in the study by Rocha, viability was evaluated using undifferentiated C17.2 cells. The activation and differentiation stimuli could make hVM1 more susceptible to arsenic toxicity. The activation of the intrinsic apoptosis pathway and simultaneous suppression of the PI3K-AKT pathway have been suggested as the mechanism of arsenic toxicity in neural stem cells [13].
On the other hand, transcription factor cAMP-Response Element Binding Protein (CREB) has been extensively studied in the nervous system and is associated with neuronal functions such as plasticity, neuronal maturation and membrane depolarization [35]. CREB regulates a wide range of genes and miRs, including miR- 132 [36]. miR-132 over-expression is induced by neurotrophins like BDNF through activation of the ERK1/2 pathway, implicating CREB phosphorylation by MSK1 [37]. Our results show high data dispersion and a trend toward downregulated miR-132 expression following arsenic exposure at 5μg/L; however, this was not statistically significant. Additionally, because CREB is an indirect miR-132 target, our results suggest an association between miR- 132 and CREB expression. Previous reports have shown that arsenic can induce alterations in CREB on the mRNA level, although this does not necessarily suggest a similar change at the protein level [38]. In addition, the expression patterns between miR-132 and CREB observed at 5μg/L of arsenic could be a mechanism of self-regulation. [38]. proposed a model for miR-132 regulation through the suppression of p300, which is a co-activator of CREB. miR-132 expression is induced due CREB activation, then mature miR-132 is able to bind to the 3’UTR of p300 mRNA and CREB activation is suppressed, allowing for miR-132 downregulation. Therefore, during the differentiation of hVM1 cells, sodium arsenite exposure may alter miR-132 expression and may be associated with the downregulation of CREB expression, suggesting that miR- 132 is a regulator in the mechanism to balance CREB recovery on the gene expression level due to p300 regulation. Several important and novel functions of miR-125b have been reported in neural differentiation, suggesting that this is a key player in the molecular cascade that contributes to the irreversible commitment of pluripotent human stem cells to the neural lineage. Actually, at least 10 targets relevant to neural differentiation and function have been identified for the action of miR-125b [39]. One of the most important targets that have been studied is nestin [40]. which has a crucial role in neural cell differentiation. A statistically significant decrease in miR-125b expression was observed in differentiating hVM1 cells exposed to arsenic in the present study. This was correlated with nestin upregulation in a dose-dependent manner following sodium arsenite exposure. The increase in the expression of nestin in differentiating cells could be a marker of damage to the differentiation and function cellular program. Our miR-125b results reveal that the neural differentiation of hVM1 cells can be disturbed in the presence of 1 and 5μg/L arsenite, although it is important verify the neuronal phenotype, which needs to be determined in further studies. In order to elucidate other possible targets for arsenic, we analyzed BDNF expression, as it promotes neural differentiation and is implicated in miR expression [41,42] However, we did not observe a statistically significant difference between untreated and exposed cultures. Arsenic had been implicated in the disruption of cellular methylation reactions, particularly in DNA methylation Several reports have shown that arsenic exposure represses DNMT mRNA expression and activity [43]. In this study, we evaluated changes in miR-148a and DNMT3b expression and found significantly augmented miR- 148 expression in hVM1 cultures exposed to 1μg/L of arsenic, the effect was the opposite at 5μg/L of arsenic as hormesis effect. Furthermore, DNMT3b expression was not significantly different compared to the unexposed group, although a trend toward downregulation in hVM1 cells exposed to both concentrations of arsenic appeared, as previously described by [13] They observed that human keratinocyte-derived HaCat cells exposed to arsenic up to 5μM for 72 h had significantly inhibited expression of DNMT1 and DNMT3a. Diminished DNMT expression could be indicated in global and gene-specific DNA hypomethylation, although the CpG methylation status not assayed. These results suggest that miR 148 is not the only regulator of DNMT3b expression, accordingly, as has been reported previously The effects of arsenic on miR expression described in this study could be an important linkage between wide physiologic alterations and a common mechanism. miRNA is considered to be a master regulator of several cellular process, including neural differentiation, hence it is possible to suggest as a likely mechanism arsenic-mediated changes to gene expression through the disruption of several signaling pathways. In agreement with our study, [44] showed that sodium arsenite affects the signaling pathways that control neural differentiation of human neural stem cells through overactivation of MEK-ERK and suppression of PI3K-AKT. Because multiple key transcription factors are affected, changes in miR and target gene expression could occur.
Conclusion
In conclusion, these findings show that sodium arsenite exposure promotes changes in miR expression, which affect important neural differentiation regulators and therefore could affect the differentiation of hVM1 cells.
References
- Kitchin KT (2001) Recent advances in arsenic carcinogenesis: Modes of action, animal model systems, and methylated arsenic metabolites. Toxicol Appl Pharmacol 172(3): 249-261.
- Chen CJ, Chen CW, Wu MM (1992) Cancer potential in liver, lung, bladder and kidney due to ingested inorganic arsenic in drinking water. Br J Cancer 66(5): 888-892.
- Rodriguez VM, Jimenez-Capdeville ME, Giordano M (2003) The effects of arsenic exposure on the nervous system. Toxicol Lett 145(1): 1-18.
- Vahidnia A, Van Der Voet, GB De, Wolff FA (2007) Arsenic neurotoxicity-A review. Hum Exp Toxicol 26(10): 823-832.
- Lindgren A, Danielsson BR, Dencker L, Vahter M (1984) Embryotoxicity of arsenite and arsenate: distribution in pregnant mice and monkeys and effects on embryonic cells in vitro. Acta PharmacolToxicol 54(4): 311-320.
- Liu S, Piao F, Peng Y, Zhong Y, Ma N, et al. (2012) Arsenic-induced inhibition of hippocampal neurogenesis and its reversibility. Neurotoxicology 33(5): 1033-1039.
- Chattopadhyay S, Bhaumik S, Chaudhury A, Das Gupta S (2002) Arsenic induced changes in growth development and apoptosis in neonatal and adult brain cells in vivo and in tissue culture. Toxicol Lett 128(1-3): 73-84.
- Ivanov VN, Hei TK (2013) Induction of apoptotic death and retardation of neuronal differentiation of human neural stem cells by sodium arsenite treatment. Exp Cell Res 319(6): 875-87.
- Kumagai Y, Sumi D (2007) Arsenic: signal transduction, transcription factor, and biotransformation involved in cellular response and toxicity. Annu Rev Pharmacol Toxicol 47: 243-262.
- Felix K, Manna SK, Wise K, Barr J, Ramesh GT, et al. (2005) Low Levels of Arsenite Activates Nuclear Factor-kB and Activator Protein-1 in Immortalized Mesencephalic Cells. J Biochem Mol Toxicol 19(2): 67-77.
- De Vizcaya-Ruiz A, Barbier O, Ruiz-Ramos R, Cebrian ME (2009) Biomarkers of oxidative stress and damage in human populations exposed to arsenic. Mutat Res 674(1): 85-92.
- Reichard, JF, Puga, A (2010) Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2(1): 87-104.
- Reichard, JF, Schnekenburger, M, Puga, A (2007) Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun 352(1): 188-192.
- Fu HY, Shen JZ, Wu Y, Shen SF, Zhou HR, et al. (2010) Arsenic trioxide inhibits DNA methyltransferase and restores expression of methylation-silenced CDKN2B/CDKN2A genes in human hematologic malignant cells Oncol Rep 24(2): 335-343.
- Xu F, Mao C, Ding Y, Rui C, Wu L, et al. (2010) Molecular and Enzimatic Profiles of Mammalian DNA Methyltransferases: Structures and Targets for Drugs. Curr Med Chem 17(33): 4052-4071.
- Martínez L, Jiménez V, García-Sepúlveda, C, Ceballos, F, Delgado JM, et al. (2011) Impact of early developmental arsenic exposure on promotor CpG-island methylation of genes involved in neuronal plasticity Neurochem Int 58(5): 574-581.
- Bartel DP (2004) MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116(2): 281-297.
- Skreka K, Schafferer S, Nat IR, Zywicki M, Salti et al. (2012) Identification of differentially expressed non-coding RNAs in embryonic stem cell neural differentiation. Nucleic Acids Res 40(13): 6001-6015.
- Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, et al. (2013) MicroRNA-based promotion of human neuronal differentiation and subtype specification. PloS One 8(3): e59011.
- Im HI, Kenny PJ (2012) MicroRNAs in neuronal function and dysfunction. Trends Neurosci 35(5): 325-334.
- Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri, N, et al. (2012) MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci 69(1): 89-102.
- Junn, E, Mouradian MM (2010) MicroRNAs in neurodegenerative disorders. Cell Cycle 9(9): 1717-1721.
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, et al. (2009) MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol, 29(19): 5290-5305.
- Cui Y, Xia Z, Han J, Sun J, Ding W, Zhao, et al. (2012) Orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci 13(1): 116.
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, et al. (2010) Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 20(4): 492-498.
- Noble EE, Billington CJ, Kotz CM, Wang C (2011) The lighter side of BDNF. Am J Physiol Regul Integr Comp Physiol 300(5): R1053-69.
- Duursma AM, Kedde M, Schrier M, Ie Sage C, et al. (2008) MiR-148 targets human DNMT3b protein coding region. RNA 14(5): 872-877.
- Sheikh MA, Malik YS, Yu H, Lai M, Wang X, et al. (2013) Epigenetic regulation of Dpp6 expression by Dnmt3b and its novel role in the inhibition of RA induced neuronal differentiation of P19 cells. PloS One 8(2): e55826.
- Wang, Z, Zhao, Y, Smith, E, Goodall, GJ, Drew, PA, et al. (2011) Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol Sci 121(1): 110-122.
- Chen T (2010) The role of MicroRNA in chemical carcinogenesis J Environ Sci Health C Environ Carcinog Ecotoxicol Rev 28(2): 89-124.
- Villa A, Liste I, Courtois ET, Seiz EG, Ramos M, et al. (2009) Generation and properties of a new human ventral mesencephalic neural stem cell line. Exp Cell Res 315(11): 1860-1874.
- Courtois, ET, Castillo, CG, Seiz, EG, Ramos, M, Bueno, C, et al. (2010) In vitro and in vivo enhanced generation of human A9 dopamine neurons from neural stem cells by Bcl-XL. J Biol Chem 285(13): 9881-9897.
- Krabbe C, Courtois, E, Jensen, P, Jørgensen, JR, Zimmer, J, et al. (2009) Enhanced dopaminergic differentiation of human neural stem cells by synergistic effect of Bcl-xL and reduced oxygen tension. J Neurochem 110(6): 1908-1920.
- O’Brien, J, Wilson, I, Orton, T, Pognan F (2000) Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267(17): 5421-5426.
- Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, et al. (2010) miR 132 regulates antiviral innate immunity through suppression of the p300 transcriptional co activator. Nat Cell Biol 12(5): 513-519.
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, et al. (2005) A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA 102(45): 16426-16431.
- Pan W, Zhu S, Yuan M, Cui H, Wang, et al. (2010) MicroRNA-21 and MicroRNA-148a contribute to DNA Hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase. 1 J Immunol 15 184(12): 6773-6781.
- De Vizcaya-Ruiz A, Barbier O, Ruiz-Ramos R, Cebrian ME (2009) Biomarkers of oxidative stress and damage in human populations exposed to arsenic. Mutat Res 674(1): 85-92.
- Karpova NN (2014) Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology 76 Pt C: 709-718.
- Rocha RA, Gimeno-Alcañiz JV, Martín-Ibañez R, Canals JM, Vélez D, et al. (2011) Arsenic and fluoride induce neural progenitor cell apoptosis. Toxicol Lett 203(3): 237-244.
- Numakawa, T, Richards, M, Adachi, N, Kishi, S, Kunugi, H, et al. (2011) MicroRNA function and neurotrophin BDNF. Neurochem Int 59(5): 551-558.
- Remenyi J, Hunter CJ, Cole C, Ando H, Impey S, et al. (2010) Regulation of the miR-212 / 132 locus by MSK1 and CREB in response to neurotrophins. Biochem J 291: 281-291.
- Wlodarczyk B, Bennett GD, Calvin JA, Craig JC, Finnell RH, et al. (1996) Arsenic-Induced Alterations in Embryonic Transcription Factor Gene Expression: Implications for Abnormal Neural Development. Dev Genet 18(4): 306-315.
- Wu J, Xie X (2006) Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol 7(9): R85.

 Research Article
Research Article