Abstract
When using a compacted version of the “I Ching” genetic code, the number of symbols is reduced to only 8 symbolic binary constants, integrated by only three symbols, being each: either the continuous or the broken horizontal line, plus the respective one word abbreviations of their resulting amino acids, namely, between four to eight accompanying the triad of lines, giving a total set of 46 possible combinations. This study is an alternate way of file compression for the genetic code, focused in the nucleotides, while another one I explored elsewhere, was focused in the groupings of amino acids, both individually and by their codon equivalents. As an Appendix 1 & 2 analogy, I add an example of “mutations” in literature, based on two early versions of “The Fair” (in its first edition of 1963, and in its first red edition of 1971), comparing its vignettes, and written in Spanish by my professor Juan José Arreola, now at his 101 year of birth.
Abbreviations: A: Amino acids; ALA: Alanine; VAL: Valine; ILE: Isoleucine; LEU: Leucine; MET: Methionine; PHE: Phenylalanine; TRP: Tryptophan; ASP: Aspartic Acid; ASN: Asparagine; GLU: Glutamic acid; GLN: Glutamine; ARG: Arginine; LYS: Lysine; SER: Serine; THR: Threonine; GLY: Glycine; PRO: Proline; HIS: Histidine; CYS: Cysteine; TYR: Tyrosine; NT: Nucleotides; U: Uracil; C: Cytosine; A: Adenine; G: Guanine; T: Thymine; N: Any Nt, R: Purines, Y: Pyrimidines; I: A, C, T (or U); H3: Triple Bonded Nucleotides in the Double Helix: C and G; H2: Double Bonded Nucleotides in the Double Helix: A and T or U.
Introduction
Previously we have seen the properties of the genetic code behaving as a computational program in the defragging [1] and the file compression [2]. Our aim here is to expand this last concept by compressing the genetic code from the point of view of the codons and not as we did before, that was by taking into account the compatibility between amino acids, for that purpose, once more as before we will use the “I Ching” as our tool for research, with which we were able to diminish the code by resting from it 40 of its codons. The advantages of this approach are that for educational purposes the students are able to see once more the enormous plasticity of the genetic code in its way to represent it, evocative as we have said before, of the same, almost limitless exchangeability in nature of all the living organisms that depend on it, but always within a clear and present natural boundary of which they are unable to jump [3], these results will also be able to help on the architecture of biological software aimed at the study and comparison of extremely long sequences.
Materials and Methods
The dimeric binary representation of the “I Ching” (also called “bigrams”) has been possible through the use of the Unicode symbols, which are, by using the font of Microsoft Ya Hei or M. Jheng Hei (but also working the next fonts: Malgun, Miryo, Gothic, Mincho UI, Ming LiU, Sim Sun, Segoe UI Symbol, etc., because if you do not have any of these, when you execute the next routine, what happens is that only a question mark within a rectangle is what appears, then you will need to select that question mark and change the font by one of the ones mentioned):
1. For the two continuous lines: 268C Alt X : ⚌ (which means
to put 268C and then to press the Alt button and then, while
pressing it, hit the “X” one);
2. For the upper broken and lower continuous line: 268E Alt
X : ⚍;
3. For the upper continuous and lower broken line: 268E Alt
X : ⚎; and finally,
4. For the two broken lines: 268F Alt X : ⚏. The method is to
consider as mentioned, the first two nucleotides, being either
made of purines or pyrimidines, or a combination of them, as
the most significant bases.
So, the equivalence of these diagrams of the “I Ching” when compared to their Yin/Yang arrows presented before [2] are as follows: Remembering that when the arrows shown in the right side of Table 1, the third dimensional spatial result is a reflection of the spiral ladders of the double helix, as shown next (Figure 1).
Table 1: Equivalences between the “I Ching” digrams (presented here) and the Yin/Yang arrows (presented in 2, there, not here, in its Appendix Figure 2; see the logic for their molecular obtaining there) as per their relation to the genetic code.
Results
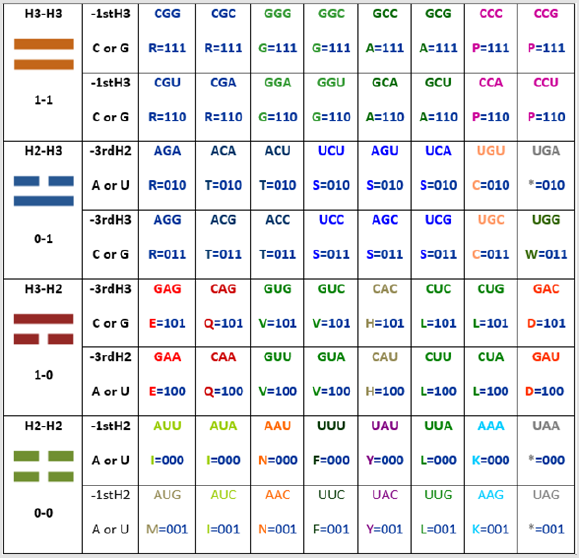
The resulting graphic is as follows (taking the first nucleotide for the first and the last comparison of codons and the third nucleotide for the second and third comparison of codons, attending to my discovery that the extreme arrows go in one direction while the center arrows go in the opposite direction [1]): These are 24 common symbols (= 384/16= 24, such as the hours of the day, twice the months of the year). Which amazingly is the same number obtained by the grouping of amino acids [2] even when the groups are different as presented here in the discussion; also, in the Appendices 1 and 2 we have the representations of the parameters of the C-rings and of the tautomerism as the other two alternatives for classification, in order to be able to integrate by using these tables the third dimensional coordinates X, Y, Z of the Cartesian graphics as demonstrated elsewhere when the 64-codon tables were used [2]. While, the resulting groupings and their representations are: The symbols belong only to the first two nucleotides (the most significant, as the third one has to do more with the regulation of transcription [3]);
First by using the ideograms of the “I Ching”, then the numbers, and then the nucleotide letters, as seen in Figure 2: one for the long line (being the molecular logic for this that the triple H bond has precisely one hydrogen more of strength when compared to the double H bond, which here is, this last one represented with the broken line), hence zero for the broken line (and that was precisely the numerical assignment given as well by Leibnitz, the first scientist to praise the “I Ching” as a binary system preceding his own binary discoveries); and finally by using the letter of the element (H) combined with the number of bonds (either three or two, in order to visually describe the physicochemical property):
For the first group (Pur-Pur_): ⚌–R , ⚌–G , ⚌–A , ⚌–P; or 1-1-R, 1-1-G, 1-1-A, 1-1-P; or H3-H3-R, H3-H3-G, H3-H3-A, H3-H3-P.
For the second group (Pyr-Pur_): ⚍–R, ⚍–T, ⚍–S, ⚍–C, ⚍–W,
⚍–* ; or 0-1-R, 0-1-T, 0-1-S, 0-1-C, 0-1-W, 0-1-*; or H2-H3-R, H2-
H3-T, H2-H3-S, H2-H3-C, H2-H3-W, H2-H3-*.
For the third group (Pur-Pyr_): ⚎–E, ⚎–Q, ⚎–V, ⚎–H, ⚎–L,
⚎–D ; or 1-0-E, 1-0-Q, 1-0-V, 1-0-H, 1-0-L, 1-0-D; or H3-H2-E, H3-
H2-Q, H3-H2-V, H3-H2-H, H3-H2-L, H3-H2-D.
For the fourth group (Pyr-Pyr_): ⚏–M, ⚏–I, ⚏–N, ⚏–F, ⚏–Y,
⚏–L,⚏–K, ⚏–* ; or 0-0-M, 0-0-I, 0-0-N, 0-0-F, 0-0-Y, 0-0-L, 0-0-K,
0-0-*; or H2-H2-M, H2-H2-I, H2-H2-N, H2-H2-F, H2-H2-Y, H2-H2-L,
H2-H2-K, H2-H2-*.
Now, when we transform each cell to its binary value, for example, for CCC or for GGG: 111, and for AAA or for UUU: 000, we have the uniform presence of the next order of binary transformed into decimal numbers per cell of each column from the top to bottom rows, we have: 7, 6, 2, 3, 5, 4, 0, 1 in decimal numbers, which are the equivalent to the next binary ones (which are shown in Figure 3): 111, 110, 010, 011, 101, 100, 000, 001; while if we multiply the hydrogen bridges per each cell as GGG being 3x3x3=27 and AAA being 2x2x2=8 and all its intermediates, we have, again going from the top rows to the bottom: 27, 18, 12, 18, 18, 12, 8, 12.
Figure 2: The H-bonds file compression of the codons by taking into account only the two first nucleotides of the codon as
Niremberg did in his initial column of his never published original handwritten drawing, until now by me [1], and here in its
original for the first time with academic purposes (see Figure 4).
Note: Here, included are the binary equivalences of A and U as 0 and C and G as 1.
Discussion
I did the representation of the complementary hexagrams of the genetic code represented by the “I Ching” in a complementary way since 2012 [4], by using the logic of the annealing of a double helix genetic code starting at the opposite extremes, being the top and the bottom, and at the left and the right, respectively, to end at the center, having obtained this representation by doing a graph of the Keto/Amino tautomerism in the Axis of the “x” (equating G and U to zero and A and C to one) when compared to the nature of the nucleotide ring, being either a purine or a pyrimidine (equating here A and G to zero and C and U to one), something that to my knowledge nobody else had done: to explain the possible logical origin of the complementary or annealing order of the “I Ching” as a genuine earliest representation of the genetic code, as far as we know.
Apart of the earlier books devoted to it, mentioned since my earlier approaches to the square tables to represent the genetic code [5], an inspiring pioneer work because of being one of the earlier works in a scientific journal, was done by Pethoukhov [6], who also obtained the table that I just mentioned, but he did it by using methods such as eigenvalues of matrices [7], and whom in his most recent reviews of the subject says things such as the next enlightening ones [8]:
1. That even Francoise Jacob said (in French) that “it may
be that the “I Ching” should be studied to capture the relationships
between heredity and language”.
2. “This ancient sequence of 64 hexagrams in Fu-Xi’s order
was identical to the ordinal series of numbers from 63 to 0 in
decimal notation” (which table corresponds to my logical and
independent molecular discovery for its ordering, as stated in [1]),
3. “By analogy, a sequence of 8 trigrams in Fu-Xi’s order is
identical to the ordinal series of numbers from 7 to 0 in decimal
notation”,
4. “If tabular cells with odd dyadic-shift numerations are
painted in black, then the well-known pattern of 64 cells of chessboard
appears in this (8х8)-matrix of dyadic shifts (this means to
alternate the first cell as white and the next as black, and so until the
end as shown in his graphic); one can think that the popularity of
many games on such chess-boards is connected with the archetypal
significance of this pattern”,
5. “Modulo-2 addition and dyadic-shifts (“notions and
operations from the field of informatics but not from physics,
chemistry, etc.”, he says) were known in Ancient Chinese culture.
They were used in constructions of Chinese tables… then the world
is created in accordance with informational principles”,
6. “The genetic coding system that has (its logic and
mathematics) a property of noise-immunity coding (another elegant
to say that it is normally resistant to mutations) at transmission of
information along a chain of generations”, and for that reason he
said: “In living matter reigns informatics of geno-logical coding”, this
also inspired by Schrodinger’s statement in 1994 that said that we
need: “to find it (¡life!): Working in a manner that cannot be reduced
to the ordinary laws of physics” (all of this said by Pethoukhov in his
live conferences, available at his webpage: http://petoukhov.com/;
otherwise, I think that his articles are somehow repetitive, except
for his recent approach of literature and its similarity to genetic
sequences, in [9].
7. Also Pethoukhov reiterates time after time that the
Chinese did not know the genetic code; in my case I agree with
that, but I understand by my biblical studies that such knowledge
was a pre-flood knowledge that after the flood was lost, that could
explain all the gigantic mammals of that time that were not chosen
to survive.
And here I wish to add a brief parenthesis to include what is known, thus far about my genome: I am Southern European in a 52%, mostly Spanish (from Catalonia), and from Andorra, and also Portuguese in a 34%; and as they learn more, they will detail even more the 18% that I have left to know in detail right now. Before I go out of Europe, the analysis of me shows that I am 3.0% broadly European (without even giving the details about this yet) and 0.5% of Ashkenazi, corrected in the Wikipedia to say that in Spain they are called Sephardic; then I am 35.3% East Asian & Native American (then the nuanced details say that out of that I am 34.9% from Northern Asian & Native American, being the 33.4% of Native American, and 1.5% is Broadly Northern Asian & Native American; interestingly, the site “23 and Me” that did my analysis says that: “Subtle linguistic affinities reveal ancient links between some Native American languages and languages still spoken in Siberia”, being a 0.2% of Broadly East Asian & Native American).
Furthermore, I have a 0.1% of Chinese Dai; of Sub-Saharan African I have 1.9% (subdivided in West African 1.6%, and from those I have 0.7% of Senegambian & Guinean, 0.1% of Nigerian, and 0.8% of Broadly West African and also 0.3% of Broadly Sub-Saharan African); and finally, I have 1.8% of Western Asian & North African (subdivided into 1.3% of North African & Arabian and 0.5 % of Broadly Western Asian & North African); and still unassigned 5.4%, and something that is called there Trace Ancestry I have less than 0.1% of Broadly Melanesian! So, this is how it looks through time the blend of my genes: Next, we will see the original handwritten table by Niremberg showing that his earliest attempt was precisely, as we have done here, to draw a table with the first two strong nucleotides as its basis (the first vertical columns or axis of the “y”).
Then we are dealing with a real medical case, the Glu210Lys mutation in the gene UBTF, plus other in MTCH2, plus other ten, expressed in excess of rRNA, biorhythms altered, loss of weight: 30 kg at 20 years of age, etc., called CONDBA (Childhood Onset Neurodegenerative Disease with Brain Atrophy). My first preliminary approach was: “GAA & GAG are for Glu; and these two are for Lys: AAA & AAG, so the change is a “G” for an “A”, or an extreme, pH speaking, of an Acid for a Basic Amino Acid”, my advice was to start feeding her with basic foods, avoiding the acid ones, intense massage, etc… here is her case: https://www.almyfoundation. org; (more recently, I was informed of an A15924G change of mitochondrial DNA to produce tRNA) so, if any other geneticist, molecular biologist or doctor wish to contact them offering their help, that will be great!
Now, when we transform each cell to its binary value, for example, for CCC or for GGG: 111, and for AAA or for UUU: 000, we have the uniform presence of the next order of binary transformed into decimal numbers per cell of each column from the top to bottom rows, we have: 7, 6, 5, 4, 2, 3, 1, 0; while if we multiply the hydrogen bridges per each cell (shown in the left side of Figure 4) as GGG being 3x3x3=27 and AAA being 2x2x2=8 and all its intermediates, we have, again going from the top rows to the bottom: 27, 18, 18, 12, 12, 18, 12, 8. And now if we multiply each result by eight as there are eight cells per row we have, for the first transformation from binary to decimal, as the total per row: 56, 48, 40, 32, 16, 24, 8, 0, why the 32 number needs to surround itself here of 32 at each side? I do not know: 32 = 24 + 8. For the second comparison of multiplying the H bonding, provides for us: 216, 144, 144, 96, 96, 144, 96, 64, and again: Why the last 144 needs to surround itself of 96 at each side: I do not know?
Furthermore, it can be said that the relation per dyad or diagram of its upper row to its lower row for the H3-H3-, H3-H2- and H2- H2- comparison is 3/2=1.5 while for the H2-H3- is 2/3=0.666... (ad infinitum). This is what I read on the internet about it: “Ratios have an inverse relationship to string length, for example stopping a string at two-thirds (2:3) its length produces a pitch one and onehalf (3:2) that of the open string”. The differences between the 24- file compression obtained by grouping the amino acids according to their common close proximity presented in [2], and the ones presented here based on the codons according to their frequency of hydrogen bonds are the next ones: In the Figure 5 we have the key for some of 3-D models of it, as it will be similar to the Fig. 13 of reference [2]. To do a backwards exercise, here you can see the transformation of the compressed genetic code represented as 12 groups reduced plus the stop codon as it can be represented in a 3-D tetrahedron way, again using the concept of the on an off switch, for the start and the end of the transcription, already presented in [10].
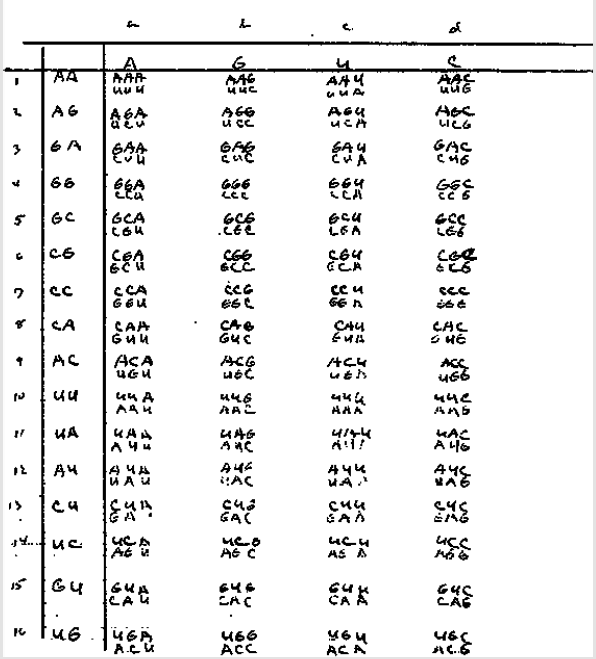
Figure 4: Original handwritten table by Niremberg showing his drive to use the first two strong nucleotides of the codons as the basis for his initial table (axis “y”), leaving the third or weak nucleotide (responsible for the speed of transcription, as seen in [4]) as the axis “x”.
Taken from: https://profiles.nlm.nih.gov/ps/access/JJBBJX.pdf Saved at: http://www.webcitation.org/75K5GUU8J Note: The table of Niremberg was also presented in an explained form in [1], where it was used as the basis for the design of synthetic chromosomes to represent the genetic code.
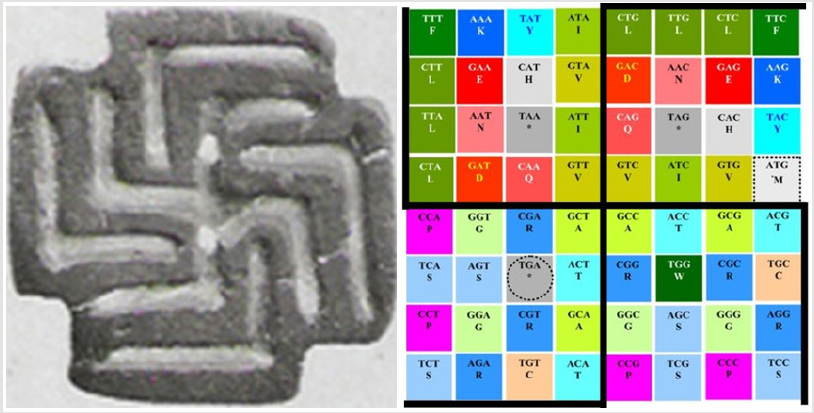
Finally, I wish to present some corrections that I just realized from my previous publications, in one of them I said “Lysine” when I wanted to say “Leucine” (Figure 5, where it says: “lysine in quadrants one and two” and it should be: “leucine in quadrants one and two”), then in the same paper [11], for the amino acid F, it wrongly says that equals CCY, when the correct equivalence is UUY (p. 719, after “Interchange between aromatic…), sorry! Then in another, I say that there were 192 codons as the sum of all the members of the RNA and of the DNA, so the multiple should be 3 x 64 = 192 (and not as it is there (2 x 64) in ref. 2 at the end of the seventh point of Negadi, where it says: “the total number of codons in DNA and RNA”); the other correction with an added figure that I want to add here is when I was talking about the bps that were paired, I forgot to multiply all of them per 3 in [1] because of the presence of the three mandatory nucleotides per codon, which means: 8 bp “times three” = 24 bp, 4 bp “times tree”= 12, etc. For example, in the next astounding discovery of the double symbol of fertility (this time with its lines drawn to ease its view):
Figure 5: Comparison of the 24 cells obtained in this work (left), by ordering the codons according to their h-bonding with a previous work showing the groupings by common amino acids (right). The only difference is that the first one separates the stop codons in two (which are all together in the other table), while the second one does the same with the serines (which are all together in the opposite table).
Note: The numerical value of multiplying the H-bonds as explained in the text is added per cell. Showing the 3/2 relation for the rows 1-1 and 1-0, while the 0-1 and the 0-0 rows have the inverse relation of: 2/3. The graphic in the right side was taken in a modified form from [10].
You can also see in Figure 7 that a clear-cut was done in [1] of all the strong codons, due to their increased stability or the harder way to separate them, with a 3H-bond in the lower part of the table compared to all the weak codons, due to their decreased stability or the easier way to separate them, with a 2H-bond in the upper part of the same. So, on the basis of that, I may suggest that the article quoted by (who also quotes my work), may put in the upper half of the circle the so-called by him “intermediate” that are in the lower half, and vice versa, in that way all the “strong” 3H codons at their center nucleotide could be together. Now, amongst some selected references quoting my works, some of the few assertive ones (as others even mistake my configuration in their reference, calling circular what is tetrahedral [12]; or guessing in an incomplete analysis (by using only 1/3 of it all, lacking to consider the properties of the C-rings and of tautomerism), at least in relation to my tetrahedron, over that molecular dictionary [13].
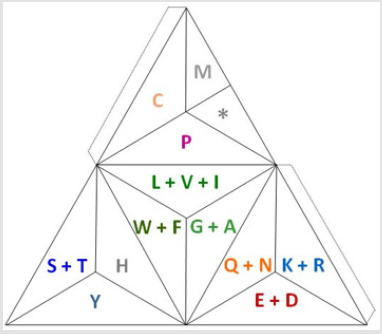
Figure 6: The most extreme file compression of amino acids in the genetic code by grouping them through their exchangeability and compatibility [11]. This is half the size of the proposal of the current article, plus the group of the stop codons acting as an off switch in the same cell as the on switch: M.
Instead of into the genes, which are the actual writings that are using the sourcebook), we have [14] who uses the figure that I developed for [15]: reaching a very nice conclusion, as they say that: “We have tried to show here that DNA polymer is not simply an inert carrier of genetic information, but rather is a dynamic partner of many other molecules in the cell, and thus capable of predetermining regulatory access to genes through its own physical chemistry”, to reach this conclusion, earlier, they also quoted my work by saying that: “We have further noticed that with the exception of only two synonymous codon sets, this apparent trend could be explained parsimoniously by a general preference for a more stable phosphate linkage between the second- and thirdbase position in the codon, again suggesting a possible selective constraint, acting outside of protein and perhaps acting on DNA flexibility via synonymous sites”.
Then [16], one of the most recent ones to-date, by quoting my work on the frequency of usage of codons in humans [4] states in an interesting way: “The most decisive codons for the (RSCU: Relative Synonymous Codon Usage frequency) algorithm were the ones, which were least commonly found in non-viral sequences. It also suggests that the frequency of usage of these particular codons is different in viral and non-viral genome, which in turn hints at different biological characteristics of viral sequences”; again, and to reach that interesting conclusion, while quoting my work they declared: “RSCU values for six (viral) codons (TCG (Ser), CGC (Arg), CGA (Arg), GCG (Ala), GTA(Val) and CCG(Pro)) were the most influential in the classification model. In the human genome, none of these 6 codons are frequently used”. The other recent one [17] quoting me in the same article [4], and in the same viral venue declared, related to my article: “Codon deoptimization of nonstructural protein genes in kRSV-DB1 (a strain of the “Respiratory Syncytial Virus” recombinant and fluorescent) was performed according to human codon usage bias. Codon usage bias in mice (Mus musculus) is similar to that in humans…”; in this important work they aim for: “The development and administration of a safe and efficacious RSV LAV (a Live-Attenuated Vaccine) to seronegative infants”, “and from indirect herd protection of other vulnerable populations” (and this speaks of the indirect spread of the effects of a vaccine, in a similar way as that of the smoke of a cigar…) (Figure 7).
Figure 7: Representation of the ancestral symbol of fertility as described in (1: Swastika on a stamp seal from the Indus culture ca. 2,800 B.C., saved at: http://www.webcitation.org/67OW4dh79, where it says: “Swastika op stempelzegel, Induscultuur ca. 2800 v.C.” (noticing that this symbol in its original form is completely independent of later political overtones, and as thus we take it), with more evidence of other cultures using the same symbol for the same purposes (in the single representation only of its outermost extremes): By the Moche and Lambayeque in South America and in Chaldea (from the 10th to the 6th centuries B.C. ) and Ilios at Troja (12th century B.C.) in the Old World, and even in the prehistoric times: http://www.webcitation.org/67Q3DxXYD).
Then, one of the impressive results, methodologically speaking, based on another of my 3-D works, in this case the 3-C culture of human cells [18] is the next one [19], declaring at the end that: “This study introduced an assay for vasoactive using magnetic 3D bioprinting. These rings structurally and functionally mimic key facets of vasoactive blood vessel segments… consistent with known vasoactive responses…”, “…to high-throughput and high-content screening and may help overcome the limitations of existing ex vivo assessments of vascular contraction. This assay will aid in the reduction of animal use in labs studying vascular biology, a key tenant of the 3Rs principles (replacement, reduction, refinement)...” And just to conclude this discussion, from other of my related collaborations, we have the next one [20], which is quoted for the incursion into the highly delicate field of “Scanning Acoustic Microscopy (SAM)”, that has no need of staining [21]; then, [22] has been recently quoted for [23], in relation to the regulation of cellular calcification.
While my most quoted first author paper is [24], which “Scopus” says that has been quoted thus far by 52 other papers, but Google Scholar says that including non-indexed documents, such as Thesis and others, the number reaches to 80), being some of the recent ones the next one [25], for another experimental innovation, where our work is supporting their “GO (Gene Ontology) Module 8”: “The enriched GO terms for Module 8 are mainly related to lipid, sterol, steroid, and cholesterol…. The authors (referring to us) showed that… absence of perilipin… produced obesity-resistant mice, adapted to this altered metabolism through upregulation of oxidative catabolic pathways and downregulation of lipid/ sterol synthetic pathways to dispose of the lipolytic products that contribute to obesity resistance. This, to some extent, shows the relations between this module and obesity. Although these experiments are conducted on mice, they may have the similar results for the humans.
Obesity is known to impair the immune function and cellmediated responses. The immune cells may infiltrate or populate in adipose tissue and promote a low-grade chronic inflammation, which represents the body’s major initial defense mechanism responding to injury or infection”. Plus, another recent one [26], saying according to my article [11]: “amino acid and codon stability can be mutually favoring genomic activities”, talking about variation in the mitochondrial diversity: “Long-range nucleotide cooccurrences have a large effect on genomic diversity. Most notably, codon motifs apparently underpinned the preferences among codon positions”; and most recently, book citations regarding my works on some of my 3D representations of the genetic code as theory [27] and on developing the methodology for the athero 3D culture of Human VSMC as practice [28].
Conclusion
It has been possible in this article to reduce the genetic code to the two first nucleotides of each of its codons, obtaining a 24 sets representation of the genetic code, similar in number to the previously demonstrated grouping of amino acids by their common codons, as it was compared here in the discussion, being even more compressed my representations previously published of the icosahedrons for the point of view of the 20 amino acids (adding the stop signs as a switch within the same cell, being this at the side of the start Met; and talking about twenty, see Appendix 3 for a related count, but this time in the work of the Mexican literature) and even before the circle with the 12 groups compatible amino acids plus the stop signals = 13! (See the details there [2]), noticing that again, it was Pethoukhov who published a comparison between the language of genetics and that of literature ([9]in his case in the Russian language, but here in the multiples of three: 294 little drawings called vignettes that are mostly on the top of each fragment of the novel “The fair”, by Arreola, and the adulteration of twenty of them by its editors).
Having concluded that many times (not all the times, but so many times) the peer-reviewers are just rats attempting to block the progress of researchers dissenting with their financial interests based on their paradigms that for them are more important than the truth, and I demonstrated this in [1] when some of them advised that my work should be forbidden from publishing, this time I send this in the way that everybody who sees my work can be its peer.
Acknowledgments
To the Creator of the Genetic Code, to my parents, and to my sister Aida for motivating the analysis of our genes; sponsored in part, also by the NIH grant: T32 HL-07812.
References
- Castro Chavez F (2012) Defragged binary I Ching genetic code chromosomes compared to Nirenberg’s and transformed into rotating 2D circles and squares and into a 3-D 100% symmetrical tetrahedron coupled to a functional one to discern start from non-start methionines through a stella octangula. JPSCB 1:1-24.
- Castro Chavez F (2014) File compression and expansion of the genetic code by the use of the Yin/Yang directions to find its phere cube. J Biodivers Bioprospect Dev 1(1): 112.
- Castro Chavez F (2012) The rules of variation expanded, implications for the research on compatible genomics. Biosemiotics p. 1-25.
- Castro Chavez F (2011b) Most used codons per amino acid and per genome in the code of man compared to other organisms according to the rotating circular genetic code. Neuro Quantology 9(4): 747-766.
- Castro Chavez F (2011) The quantum workings of the rotating 64-grid Genetic Code. Neuro Quantology 9(4): 728-746.
- Petoukhov SV (1999) Genetic code and the ancient Chinese book of changes. Symmetry Cult Sci 10(3-4): 211-226.
- Petoukhov SV (2016) The system-resonance approach in modeling genetic structures. BioSystems 139: 1-11.
- Petoukhov SV (2016) The genetic code, 8-dimensional hypercomplex numbers and dyadic shifts.
- Pethoukhov SV (2018) Structural Connections between Long Genetic and Literary Texts.
- Castro Chavez F (2016) Anatomical Mnemonics of The Genetic Code: A Functional Icosahedron and the Vigesimal System of the Maya to Represent the Twenty Proteinogenic Amino Acids. J Biol Nat 5(3): 140-147.
- Castro Chavez F (2010) The rules of variation: amino acid exchange according to the rotating circular genetic code. J Theor Biol 264(3): 711-721.
- Anton A Komar (2016) The “periodic table” of the genetic code: A new way to look at the code and the decoding process. Translation (Austin) 4(2): e1234431.
- Hervé Seligmann, Ganesh Warthi (2017) Genetic Code Optimization for Cotranslational Protein Folding: Codon Directional Asymmetry Correlates with Antiparallel Betasheets, tRNA Synthetase Classes. Comput Struct Biotechnol J 15: 412-424.
- Babbitt GA, Alawad MA, Schulze KV, Hudson AO (2014) Synonymous codon bias and functional constraint on GC3-related DNA backbone dynamics in the prokaryotic nucleoid. Nucleic Acids Res 42(17): 10915-10926.
- Castro Chavez F (2012) A tetrahedral representation of the genetic code emphasizing aspects of symmetry. Bio-Complexity 2: 1-6.
- Bzhalava Z, Tampuu A, Bała P, Vicente R, Dillner J (2018) Machine Learning for detection of viral sequences in human metagenomic datasets. BMC Bioinformatics 19(1): 336.
- Rostad CA, Stobart CC, Todd SO, Molina SA, Lee S, et al. (2018) Enhancing the Thermostability and Immunogenicity of a Respiratory Syncytial Virus (RSV) Live-Attenuated Vaccine by Incorporating Unique RSV Line19F Protein Residues. J Virol 92(6): e01568-e1617.
- Castro Chavez F, Vickers KC, Lee JS, Tung CH, Morrisett JD (2013) Effect of lyso-phosphatidylcholine and Schnurri-3 on osteogenic transdifferentiation of vascular smooth muscle cells to calcifying vascular cells in 3D culture. Biochim Biophys Acta 1830(6): 3828-3834.
- Tseng H, Gage JA, Haisler WL, Neeley SK, Shen T, et al. (2016) A high-throughput in vitro ring assay for vasoactivity using magnetic 3D bioprinting. Sci Rep 6: 30640.
- Ghosn MG, Mashiatulla M, Mohamed MA, Syed S, Castro Chavez F, et al. (2011) Time dependent changes in aortic tissue during cold storage in physiological solution. Biochim Biophys Acta 1810(5): 555-560.
- Riaz Akhtar, J Kennedy Cruickshank, Xuegen Zhao, Brian Derby, Thomas Weber (2016) A pilot study of scanning acoustic microscopy as a tool for measuring arterial stiffness in aortic biopsies. Artery Res 13: 1-5.
- Vickers KC, Castro Chavez F, Morrisett JD (2010) Lyso-phosphatidylcholine induces osteogenic gene expression and phenotype in vascular smooth muscle cells. Atherosclerosis 211(1): 122-129.
- Tamer Sallam, Henry Cheng, Linda L Demer, Yin Tintut (2013) Regulatory Circuits Controlling Vascular Cell Calcification. Cell Mol Life Sci 70(17): 3187-3197.
- Castro Chavez F, Yechoor VK, Saha PK, Martinez Botas J, Wooten EC, et al. (2003) Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in perilipin knockout mice: a microarray gene expression profile. Diabetes 52(11): 2666-2674.
- Zhang S, Zhao H, Ng MK (2015) Functional Module Analysis for Gene Coexpression Networks with Network Integration. IEEE/ACM Trans Comput Biol Bioinform 12(5): 1146-1160.
- Shinde P, Sarkar C, Jalan S (2018) Codon based co-occurrence network motifs in human mitochondria. Sci Rep 8(1): 3060.
- Stcherbic VV, Buchatsky LP (2016) Living Matter: Algebra of Molecules. Florida: CRC Press pp. 160.
- Tseng H, Raphael RM, Killian TC, Souza GR (2018) Magnetic Nanoparticles for 3D Cell Culture. (ch.10 In: Dobson J, Rinaldi C (Eds.), Nanomagnetic Actuation in Biomedicine: Basic Principles and Applications. CRC Press. Taylor & Francis, London, pp. 229-244.

 Research Article
Research Article