Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Alaverdian D1, Fedyakov M1, Polennikova E4, Ivashchenko T3, Sarana A1,2, Shcherbak S1,2, Urasov S1, Makarenko S1 and Glotov O*1-3
Received: February 21, 2019; Published: February 28, 2019
*Corresponding author: Oleg Glotov, Saint Petersburg City Hospital No.40, Russia, 197706, Saint Petersburg, Borisova street, 9, Russia
Oleg Glotov, Saint Petersburg State University, Russian Federation, 199034, Saint Petersburg, Universitetskaya emb., 7/9, Russia
Oleg Glotov, D. O. Ott Research Institute of Obstetrics and Gynecology, Russia, 199034, Saint-Petersburg, Mendeleevskaya line, 3, Russia
DOI: 10.26717/BJSTR.2019.15.002693
According to a modern classification, there are two forms of inherited ichthyosises: syndromic and nonsyndromic, each of them consists of more than ten different nosologies [1]. More common types of the ichthyosis are X-linked recessive (prevalence 1/2000- 6000 in men) and autosomal dominant, or ichthyosis vulgaris with incomplete penetrance (1/250-1000) [2,3]. The X-linked form is associated with mutations in steroid sulfatase gene STS noteworthy there is a full deletion of the gene in 90% of cases [4-6]. Ichthyosis vulgaris is caused by heterozygous mutations in the gene FLG encoding filaggrin [3]. It is important to note that clinical forms of these diseases are indistinguishable. The aim of this study was the searching for pathogenic or likely pathogenic mutations which associated with various forms of the inherited ichthyosis such as other inherited diseases with the similar phenotypic signs. The sequencing was held on HiSeq 4000 sequencer (“Illumina”) by paired-end reading (2x150 bp). For the processing of the probes we used the method of selective capture of the coding DNA sequences “MedExome”[7]. The identified mutation p.Arg2037Ter in heterozygous condition was described before as pathogenic in data bases [9]. Also there was no coverage of STS gene sequence, which means that the patient has a full deletion of STS gene. Was found that the patient carries two pathogenic mutations which are related to different forms of the inherited ichthyosis. For the genetic counseling such information might be very valuable because of the clinical manifestation similarity. It is recommended to analyze both of genes STS and FLG to exclude the combined forms of ichthyosis.
Keywords: Inherited Diseases; Skin Diseases; Ichthyosis; NGS
Inherited cornification diseases (ichthyosises) make up a large group of mendelian disorders which vary clinically and etiologically. In general, all of them lead to the common feature – total or partial skin lesions which are caused by exfoliation or hyperkeratosis [1]. According to a modern classification, there are two forms of inherited ichthyosises: syndromic and nonsyndromic, each of them consists of more than ten different nosologies [1]. More common types of the ichthyosis are X-linked recessive (prevalence 1/2000- 6000 in men) and autosomal dominant, or ichthyosis vulgaris with incomplete penetrance (1/250-1000) [2,3]. The X-linked form is associated with mutations in steroid sulfatase gene STS noteworthy there is a full deletion of the gene in 90% of cases [4-6]. Ichthyosis vulgaris is caused by heterozygous mutations in the gene FLG encoding filaggrin [3]. It is important to note that clinical forms of these diseases are indistinguishable [10]. The presented clinical case points out the necessity of molecular genetic testing even if the clinical diagnosis is clear.
A 40-year-old man was referred to our department for the genetic counseling. He had complaints to dry and flaky skin and had been suffering from ichthyosis since birth – skin problems had begun in the first month of life. The skin was covered by the dark scales. The treatment had been selecting empirically and there wasn’t any stable effect. The biopsy had taken place when he was 17 years old and indicated an increased level of cholesterine-3 sulfatase which led to diagnosis X-linked ichthyosis. The selected therapy consisted of a course taking vitamin A, a cream usage and an UV-based therapy and led to the improvement over the time. By the studying the family history it was revealed that the father was healthy, but the mother and the sister had been having problems with dry skin. At the moment of the checkup obvious symptoms hadn’t observed, although some dryness of skin covering had noticed mostly in the areas of the face and external hips surfaces (Figures 1 & 2), the nail plates was without any deformation or changes. It was considered to do the genetic analysis in order to verify the diagnosis and to compose the progeny forecast. We analyzed the patient DNA and two healthy donors (male and female) DNAs as controls.
DNA extraction from WBC was carried out by sorbent method using the standard protocol. At first, we were searching for pathogenic or likely pathogenic mutations which are associated with various forms of the inherited ichthyosis such as other inherited diseases with some similar phenotypic signs. The sequencing was held on HiSeq 4000 sequencer (“Illumina”) by paired-end reading (2x150 bp). For the processing of the probes we used the method of selective capture of the coding DNA sequences “MedExome” [7]. Bioinformatical calling and filtering of results was carried out with GATK software according to the BROAD Institute recommendations. For the annotation of the founded variants we used reference sequence RefSeq, standard nomenclature HGVS and our self-developed algorithm [8]. Classification of the found variants was presented according to recommendations of American College of Medical Genetics [9] as well as literature data. The verifying the full deletion of STS gene was carried out with our self-invented multiplex PCR system with the specific primers to 1 and 10 exons to STS gene and to APOC3 gene (as an amplification control).
The NGS showed no coverage of the coding and flanking sequences of STS gene (Figure 3). We verified 1 and 10 exons deletion by PCR-RFLP (Figure 4). Additionally, we found out a heterozygous variant of FLG gene (rs200002200) which leads to nonsense mutation in 2037 position (p.Arg2037Ter: c.6109C>T) (Figure 5).
Figure 3: S – sample, C- control. Black arrows show the absence of coverage in coding sequence of STS gene.
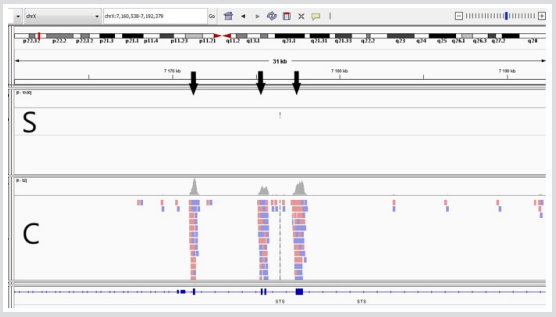
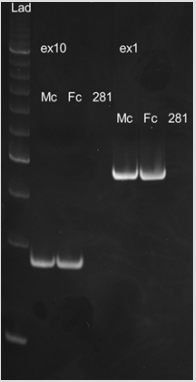
Figure 4: The absence of the amplification product in patient sample.
Lad – Ladder, Мс – Healthy male, Fс – Healthy female, 281 – Patient, ex1 – Exon 1, ex10 – Exon 10.
STS gene is located on the X chromosome at band Xp22.3 and encodes steroid sulfatase. These gene mutations are caused the development of X-linked recessive ichthyosis (OMIM:308100), wherein there is a full STS deletion in up to 90% of cases. Inasmuch as there was no coverage of STS gene sequence, the patient has a full deletion of STS gene. FLG gene encodes profilaggrin which converts to filaggrin – a protein playing a vital role in the skin structure. The mutations in FLG gene lead to ichthyosis vulgaris (OMIM: 146700). The identified mutation p.Arg2037Ter in heterozygous condition was described before as pathogenic in data bases [10]. The penetrance of this mutation is reduced and amounts to 90%. This amino acid substitution causes to the deficient protein synthesis and also is a nonsense mutation. Thus, the patient carries two pathogenic mutations which are related to different forms of the inherited ichthyosis. For the genetic counseling such information might be very valuable because of the clinical signs similarity. It is recommended to analyze both of genes STS and FLG to exclude the combined forms of ichthyosis.
This study was funded by Russian Science Foundation grant № 14-50-00069.


