Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Pascal Pedini1,2, Noura Kouba1, Matthieu Riquier1, Sophie Simon1, Agnès Basire1, Frédéric Fina3,4, Claire Galambrun5, Jacques Chiaroni2 and Christophe Picard*1,2
Received: January 11, 2019; Published: January 24, 2019
*Corresponding author: Christophe Picard, Laboratoire d’Immunogénétique, EFS Alpes Méditerranée, 149, Boulevard Baille, 13005 Marseille, France
DOI: 10.26717/BJSTR.2019.13.002420
Allogeneic hematopoietic stem cell transplantation (allo-SCT) is today an established cure for many malignant and non-malignant hematological disorders. The post-allo-SCT monitoring is very important in the engraftment phase to detect graft failure, but also in the following of post-SCT to detect the risk of disease relapse in malignant disease. The quantification of chimerism is the determination of the proportion of donor cells in recipient blood after allo-SCT. Currently, two complementary molecular biology techniques, STR PCR and qPCR methods, are routinely performed in laboratories. The practical principle is, after to have obtained informative marker between recipient and donor, the chimerism of donor/recipient is quantified by the informative markers.
The STR method is based on amplification of 16-18 STR loci, which are short base sequences on chromosomes distributed throughout the genome. Each STR marker is a set (or system) of many alleles, differing in the number of tandem repeats of this sequence. After their amplification, the separation of STR products and fluorescence detection are performed on a capillary electrophoresis instrument. The calculation of amount of donor or recipient chimerism is based on ratio of informative donor and recipient signals [1]. The real-time PCR (qPCR) using Taqman technology has been developed for the chimerism analysis using insertion/deletion polymorphisms [2]. 30 to 50 probes are identified by the manufacturers. The chimerism assay qPCR protocol includes amplifications of donor -specific allele, recipient specific alleles and active reference on DNA samples of post-SCT and donor and recipient pre-SCT.
The relative quantification of recipient chimerism is calculated in function of donor and recipient pre-SCT values. These two techniques have been showed accurate and reproducible. However, their use is mainly limited by their range of analysis, ie 80% - 5% for STR, 30% -0.1% for RQ-PCR [3-5], which requires managing two techniques within the laboratories. Indeed, STR assay is better method than qPCR to detect mixed chimerism after SCT for hemoglobinopathy whereas qPCR is better to detect the minimal residual disease after SCT for leukemia, because STR method lacks sensitivity. Furthermore, the recent development of micro-transplantations (chimerism < 1%) requires a chimerism technology with a very low sensitivity. Thus, a chimerism technique that combines both a large range of analysis and very low sensitivity would be very interesting. So, the digital droplet PCR (ddPCR) could be a good alternative to these issues.
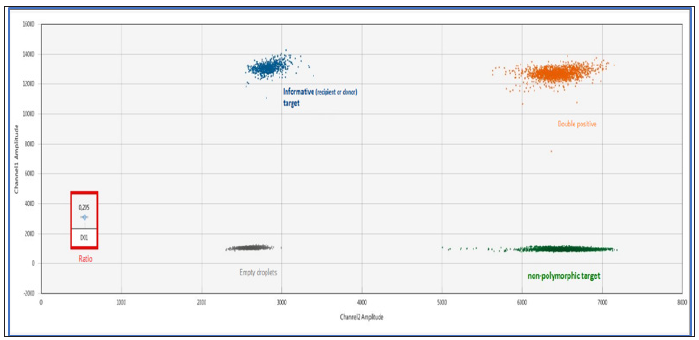
Digital droplet PCR (ddPCR) is a third generation technology that complements traditional end-point PCR and real-time PCR. It was developed to overcome few limitations of conventional amplification techniques, for the detection of small amounts of nucleic acids. This technology is in a full swing because of its high sensitivity and major applications in various domains such as oncology. With the ddPCR, the samples are partitioned into oils droplets so that each droplet becomes a separate sample with 0, 1 or more copies of the target DNA molecule. The droplets are thermally cycled to end-point and each droplet is read to determine the fraction of positive droplets. Using Poisson statistics, the absolute number of starting copies can very accurately be determined. By using TaqMan probes corresponding to two different targets, the relative proportion of each target can be determining. The system of quantification of chimerism by ddPCR is shown in Figure 1.
Note: The informative “recipient” or “donor” marker has a FAM (blue cloud) fluorescence and the marker targeting a nonpolymorphic gene has a HEX fluorescence (green cloud). Quantification of donor or recipient chimerism is obtained by the ratio (red frame): informative target / non-polymorphic target.
Figure 1:Quantification of the chimerism by ddPCR using the Biorad QX200 droplet reader (example of 20% donor chimerism).
Several teams have studied the analytical performance of quantification of chimerism by ddPCR [3]–[8] using principally probes / primers targeting a region of the Y chromosome, the AMELY gene or the SRY gene. As early as 2013, George et al. [4] show a linearity of the quantification of chimerism by ddPCR, with a correlation coefficient of R = 0.9999 for a measurement range of 0.10% to 50%. In 2016, Santurtun et al. [6] showed a strong correlation between quantification of chimerism by ddPCR and STR. This is confirmed by Stahl et al. [3] who also found an excellent correlation between the ddPCR technique and STR (R> 0.98). The sensitivity of the ddPCR were evaluated to <0.1%. Then, in 2017, Waterhouse et al. [5] showed a linear quantification of chimerism over a range of 0% to 50% with a Y-linked probe (R² = 0.9946) and an X-linked probe (ABCD1) (R² = 0.9416), a correlation between the ddPCR technique with the STR technique (R² = 0.920) and established a method sensitivity at 0.01%. The two most recent studies (2018) [7,8] confirm the findings of previous studies by confirming a good linearity, sensitivity and strong correlation of ddCPR with STR and FISH techniques.
Waterhouse et al. [5] investigated the potential predictive character of quantification of chimerism by ddPCR for relapse diagnosis in 32 patients relapsing after allo-SCT. In 15 of these patients (48%), the increase of recipient chimerism was detected earlier by ddPCR than by STR. The average detection of an increase of recipient chimerism is 65 days and 155 days before the relapse diagnosis for STR and ddPCR, respectively. The authors conclude that quantification of recipient chimerism by ddPCR is predictive for relapse diagnosis after allo-SCT, with an average difference of 90 days between ddPCR and STR. Another team showed that the combination of MRD and chimerism markers in a ddpcr platform was a practical, sensitive, and accurate diagnosis tool for comprehensive assessment of the molecular remission status of recipient post-SCT [9].
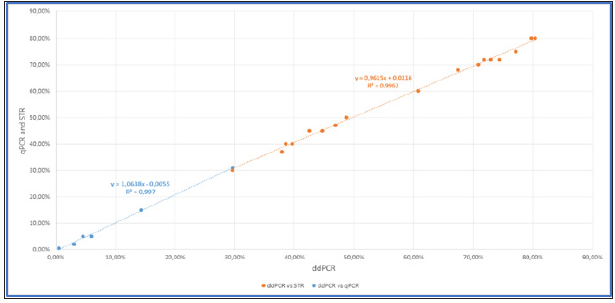
Our team has also evaluated the ddPCR for the quantification of the chimerism after allo-SCT using homemade Y-linked probe (SO2) or autosomal probe (SO8b) [2]. Our results allow us to agree with the literature and for patients with low chimerism (ddPCR is more sensitive than STR) and for patients with mixed chimerism (accuracy of ddPCR is very more important than qPCR). We confirmed that the quantification of chimerism by ddPCR is reliable over a wide range of measurement (from 0.05% to 100%) (Figure 2a). The limit of detection is < 0, 01%, compatible to microchimerism detection. We also showed a strong correlation of ddPCR with qPCR and STR techniques in their respective measurement interval (qPCR from 1% to 30%, STR from 30% to 100%) (Figure 2b). Thus, the correlation to current qPCR and STR techniques and the linearity of measurement make the ddPCR technique as a unique method for the quantification of chimerism.
Figure 2a: Linearity of chimerism quantify by ddPCR (blue: Y-linked probe (SO2); orange: autosomal probe (SO8b)). A serial dilution of mixing “man in woman” DNA (100%, 70%, 50%, 20%, 1%, 0.5%, 0,1%, 0.05%) were tested by the ddPCR technique.
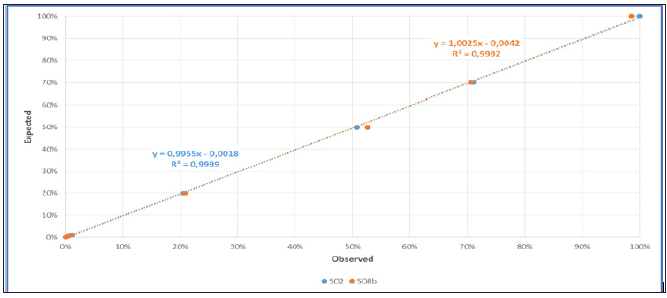
Figure 2b: Correlation of percentage of chimerism between ddPCR and qPCR /STR (blue: qPCR; orange: STR). Twenty-three samples from 4 patients (post allo-SCT) have been tested.
We adapted a technique with probes used in qPCR, but we also validated that all chimerism qPCR commercial kits are adaptable to ddPCR (data not shown). Furthermore, recently, ddPCR commercial kits marked CE-IVD, have been developed by two manufacturers (kit Quimera dpcr, Imegen® or kit Mentype Chimera, Biotype®). Our limited tests suggested that these kits are accurate and reproducible with a similar sensitivity (0.05%) (data not shown). In conclusion, the detection and quantification of chimerism by ddPCR is a sensitive, accurate, simple and inexpensive method (i.e no need for standard curves, efficiency controls, and multiple replicates as qPCR) for monitoring SCT patients. Additionally, the method can be largely automated. Furthermore, this technique seems also to be very interesting to detect graft-derived cell-free DNA (GcfDNA) in organ health recipients, since higher GcfDNA levels are indicative of acute rejection.


