Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Nawres N Jaber*
Received: November 30, 2018; Published: January 18, 2019
*Corresponding author: Nawres N Jaber, Department of Microbiology, College of Veterinary Medicine, Iraq
DOI: 10.26717/BJSTR.2019.13.002382
Two strains of Clostridium isolated from soilnear the gas stations in Basrah, Iraq, was identified by 16S rRNA sequencing as Clostridium bifermentans strain 0910-0608 and - Clostridium bifermentans strain 0912-02001 Polyhydroxyalkanoates (PHA) production by these strains were investigated. The extracted PHA was characterized by FTIR spectroscopy. It was found that these strains are PHA producers.
Keywords: PHA; Clostridium Bifermentans; Soil Sampels
In recent years, there has been a shift in public opinion with people becoming more ecologically aware. The shift in public opinion has driven industries to investigate biodegradable alternatives to plastic which are not manufactured using petrochemical methods [1,2]. Poly hydroxyl alkanoates (PHAs) are among the top group of biopolymers that have been intensively investigated and commercialized. PHAs produced by a broad range of microorganism. PHA can be synthesized by over 30 % of soil inhabiting bacteria [3]. Accumulating PHAs is a natural way for bacteria to store carbon and energy, when nutrient supplies are imbalanced. These polyesters are accumulated when bacterial growth is limited by depletion of nitrogen, phosphorous [4] or oxygen and an excess amount of a carbon source is still present. As PHAs are insoluble in water, the polymers are accumulated in intracellular granules inside the cells. It is advantageous for bacteria to store excess nutrients inside their cells, especially as their general physiological fitness is not affected. By polymerizing soluble intermediates into insoluble molecules, the cell does not undergo alterations of its osmotic state. Thus, leakage of these valuable compounds out of the cell is prevented and the nutrient stores will remain securely available at a low maintenance cost [5].
Samples were collected from the area near the gas stations in Basrah city. Soil samples 5cm deep from surface was used for isolation of the bacteria. About 15-20g of soil samples scraped with sterile plastic jars, and for the isolation of organisms one gram of soil sample is dispensed in 10ml of sterile distilled water. This is mixed vigorously and 0.1ml were pre-cultured on modified PY medium according to [6] the pre- cultivation step was conducted under anaerobic condition using Anaerobic Jar at 30oC for 7days. After incubation period the isolates were studied for morphological and biochemical characteristics. Initially, spread onto blood agare and egg yolk agar plates . The plates were incubated for 24h under anaerobic condition using Anaerobic Jar at the temperature of 30oC. then Gram staining was performed after which biochemical characterization was done to identify the phenotypic characters of the bacterial isolates, indol production , voges- proskauer and citrate utilization.
Colonies were cultured on nutrient agar plates enriched with 1% glucose. The plates were incubated at 300C for 72hrs under anaerobic condition using Anaerobic. Then, the culture plates were flooded with 0.02% alcoholic Sudan black B to stain bacterial colonies followed the method of [7]. The plates were kept undisturbed for 30min. The excess dye was then decanted , rinse with 100% ethanol. PHA producing colonies appeared bluish black. Isolates that show positive reaction with Sudan black B were checked for PHA production by using specific stain , Nile blue A stain. Bacterial isolates were grown on a modified solid HM medium with final concentration of 0.5μg dye per mL of the medium. Petri dishes were incubated at 30oC for 4 days under anaerobic condition using Anaerobic Jar. The agar plates were then exposed to ultraviolet light (312nm) to detect the presence of intracellular PHA granules in the bacteria. The colonies with fluorescent bright orange were chosen for further studies [8].
Genomic DNA Extraction: The DNA of the isolates which show positive results for PHA production was extracted by genomic DNA purification kit. The result was detected by electrophoresis on 0.8 % Agarose and showed under UV light in which the DNA appears as bands [9].
Identification by Polymerase Chain Reaction (PCR): Primers for 16SrDNA F- 5’-AGAGTTTG ATCCTGGC-3’ and R- 5’-GGTTACCTTGTTACGACTT-3’are used [10] along with PCR program selected from adapted by [11,12]. The amplification steps abstracted from [13] with little changes.
Purification, Sequencing and Manipulation of Data For PCR Product: The products of PCR were purified and sequenced at Macrogen company laboratories, Korea for complete identification of bacterial isolates.
PHA Production and Extraction in Liquid Culture: A culture medium of the same previous ingredients except agar was used. The flasks containing 100ml of liquid medium were inoculated with 0.1ml of 1 O.D at 540 nm of the bacterial suspension and incubated un aerobically at 300C, for 7 days. Cells were harvested by centrifuging. PHA was extracted using sodium hypochlorite method by [14].
Effect of Initial Ph and Nacl Concentration On PHA Production: Different PH values (2, 4, 7, 9 ,11) and NaCl concentrations ( 0.5, 1, 3, 5, 9, 12 ) were tested in order to detected there effect on PHA production from the isolated strains [15].
Fourier Transform Infrared Spectroscopy Analysis(FTIR): FTIR analysis of the polymer that extracted from this bacterial strain was carried out by Jasco FTIR 4200 spectroscopy, Japan. in IR range of 4000-400 cm-1. Peak values obtained were analysed to interpret the presence of specific functional groups in the extracted polymer [16].
PY medium proposed by Biebl and Spöer (2002) was tested for its ability to isolate anaerobic bacteria from natural samples. The aim of this work was to isolate bacterial strains that is able to produce PHA. Accordingly, bacteria was isolated from gas station soil samples. Bacterial colonies appeared with a creamy color with beta-hemolytic on blood agar. Growth on egg yolk agar also showed a significant effect on the production of lecithinase, positive for indol production negative for indole production and negative for voges- proskauer and citrate utilization , Staining has shown that isolates are Gram positive rods
Colonies of the two present isolates showed positive reaction for Sudan black B stain . The colonies stained bluish black with high color intensity as shown in Figure 1 . The colonies of these isolates showed bright fluorescence on HM agar plates containing the specific Nile blue dye when exposed to uv light (Figure 2). The bright color turn bright orange. In the present study, this confirm that the isolates are definitely PHA producer
Figure 1: Colonies on 1% glucose enriched nutrient agar showing positive reaction with Sudan black B stain.
Genomic DNA Extraction and Detection: Electrophoresis technique has revealed clear isolated DNA for the isolate
Amplifying of the 16S rDNA Gene by (PCR) Technique: By using a universal primer, results have obtained the required band of 16S rDNA for the isolate along with electrophoresed ladder in the region of 1500bp, as in the (Figure 3). The results of 16S rDNA nucleotides sequencing for the isolates were presented and the isolates were identified to the level of strain, the isolates had an identity of 100 % with strains of Clostridium bifermentans strain 0910-0608 and - Clostridium bifermentans strain 0912-02001. The Gene sequencing was shown in Figures 4 & 5.
Figure 3:Amplification of 16S rDNA in the region 1500bp Sequencing for 16S rDNA and identification of bacterial species.

Figure 4: Gene sequencing of Clostridium bifermentans strain 0910-0608 isolated in the present study.

Table 1: PHA production values and residual biomass Effect of initial pH and NaCl concentration on PHA production.
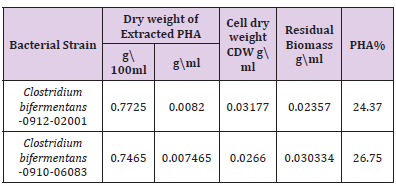
PHA production and Extraction in Liquid Culture: The PHA produced by the bacterial strains was extracted by lysing the cells with Sodium hypochlorite and chloroform as a solvent for the extraction. The residual biomass was estimated as difference between the dry cell weight and dry extract of PHA, the percentage of intracellular PHA accumulation was estimated as the percentage composition of PHA present in the dry cell weight (Table 1).
Effect of Initial PH and NaCl Concentration on PHA Production: The maximum yield of PHA production was determined at PH 7, while the production was reduced at alkaline and acidic PH values by using different concentration of NaCl, the maximum yield of production was done at (3,5) of NaCl (Figures 5 & 6).
Figure 5: Gene sequencing of Clostridium bifermentans strain 0912-02001isolated in the present study.

Fourier Transform Infrared Spectroscopy Analysis(FTIR): Powdered PHA were analyzed by FTIR to determine the functional groups of PHA . In the present study FTIR spectroscopy was performed between frequency ranges of 4000- 400 cm-1. The polymers extracted showed the intense absorption at 3425 and 3293.34 cm-1 for ( O-H ) bending group, other absorption bands at 2960,68 and 2934 cm-1 for (C-H) stretching group. IR spectra of the polymers revealed the presence of marked peaks at wave numbers 1634 and 1648.84 cm-1 representing the presence thio ester carbonyl ( C=O ) stretching groups , the marked peaks at wave numbers of 1734 due to the presence of ( C=O ) ester stretching groups . Other absorption bands at 1443, 1400, 1453 cm-1 were observed for ( CH3 ) group (Figures 7-9).
Today, most research efforts concentrate on the isolation of PHA producing microorganisms from different sources and improvement of PHA producing abilities of microorganisms [17]. In the present study, bacterial isolates were obtained from soil near the gas stations in Basrah city. For the rapid detection and isolation of PHA producing bacteria Sudan black B and Nile blue A staining viable colony methods were used. The isolates showed black - blue coloration when stained with Sudan black B, preliminary screening agent for lipophilic compounds and these isolates showed positive results with Nile blue A staining specific dye for the PHA granules [18] used these methods to screen the potential PHA producing bacteria from soil. By applying PCR technique an amplified region was shown in 1500 bp approximately which was in agreement with the result of [12], because the 16S rDNA appears in this region in all bacteria. The FT-IR technique was used in this study with spectra range of 4000 - 400 cm-1 several methods have been reported for the qualitative analysis of PHA, including GC, NMR etc. but FT-IR was reported to be the most rapid and simple method [19] it not require complicated sample preparation and can be used to detect the extracted PHA as well as intracellular PHA in the dried cells.


