Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Nathaniel Elkaim and Radhashree Maitra*
Received: December 23, 2018; Published: January 09, 2019
*Corresponding author: Radhashree Maitra, Department of Biology, Yeshiva University New York, USA
DOI: 10.26717/BJSTR.2019.12.002328
The immune system is commonly divided into two broad sections: innate immunity and adaptive immunity. Innate immunity is described as non-specific and hereditary whereas adaptive immunity is characterized by high specificity encoded somatically [1-5]. The adaptive immune system has two functional cell types: T cells and B cells [6-10]. While B cells participate in humoral immunity by releasing antibodies into the body to opsonize or incapacitate foreign bodies, cytotoxic T cells attack individual cells that may be compromised. This system of developing immunity against a specific antigen is dependent on antigen presentation to T cells [11].
The primary molecule of antigen presentation is the major histocompatibility complex (MHC) which are loaded with short peptides [14]. While MHCII presents peptides from extracellular pathogens to CD4+ helper T cells, MHCI is presents peptides from intracellular pathogens to CD8+ cytotoxic T cells [15]. T cells bind to MHC and antigen via T cell receptors (TCRs), highly specific receptors refined somatically by VJ recombination, very similar to antibodies [1] (p. 115). The structure of TCR is similar to the Fab fragment of an antibody, consisting of two chains, α and β [1] (p.114). MHC contains a short, shallow grooved on which it presents a peptide [16]. All antigens go through antigen processing before presentation on MHC, a process in which foreign proteins are processed into smaller peptides [17]. Proteins can be degraded, cut, and trimmed to size before being presented on MHC. MHCI, specifically, usually presents nanomers. As a result, MHC is capable of presenting any protein. Contributing to the diversity of antigens recognized by MHC are the array of subclasses therein and polymorphism said sublclasses, many of which are highly polymorphic [18].
While MHC efficiently presets protein antigens, other forms of antigens exist. With regards to microbial pathogens, specifically, the presentation of lipid antigens can be crucial in generating an effective immune response [1] (p. 314-5). Presentation of lipids to T cells is accomplished via MHCI related proteins CD1 [19] and MR1 [20], which contain an α chain and non-covalently associated β2-microglobulin. The understanding of alternate presentation systems has developed slowly. The first paper describing CD1-restricted T cells was published in 1989, but the nature of the antigen presented was not precisely identified. The emergence of lipids as T cell antigens presented by CD1 molecules was only established five years later by the discovery of the antigenic properties of mycolic acid. Currently, a variety of lipids, from both self- or non-self-origin, are known to bind CD1 molecules and to participate in lipid-specific T cell development and activation” [5]. While MHC I and II research followed a disease-to-molecules trajectory, CD1 research is unfolding as a molecules-to-disease story” [7], as evident from the slow discovery of its ligands and associated cells in the 1990’s and crystal structure through the early 2000s. The recent advent of CD1 tetramers has allowed progress of research into novel ligands and helped gain an understanding the mechanism of the binding of CD1 to its ligands, and the specificity of TCRs for CD1-ligand complexes [8].
The CD1 genes are located on chromosome 1, are non-polymorphic, and contain only five isotypes: CD1a-e [21]. CD1a-c are categorized together as Group I CD1 molecules, CD1d comprises Group II, and CD1e comprises Group III. The isotypes vary in localization and function. Group I CD1 molecules are almost exclusively expressed on thymocytes and dendritic cells (DCs) and are present in humans but not in rodents. “CD1d has a wide expression pattern and is present in both hematopoietic and nonhematopoietic derived cells. CD1e is expressed on DCs but does not function as an antigen presenting molecule, since it is not present at the plasma membrane. This molecule functions as a lipid transfer protein (LTP). CD1e promotes the loading and unloading of lipids into CD1d and also influences lipid presentation by CD1b and CD1c” [5].
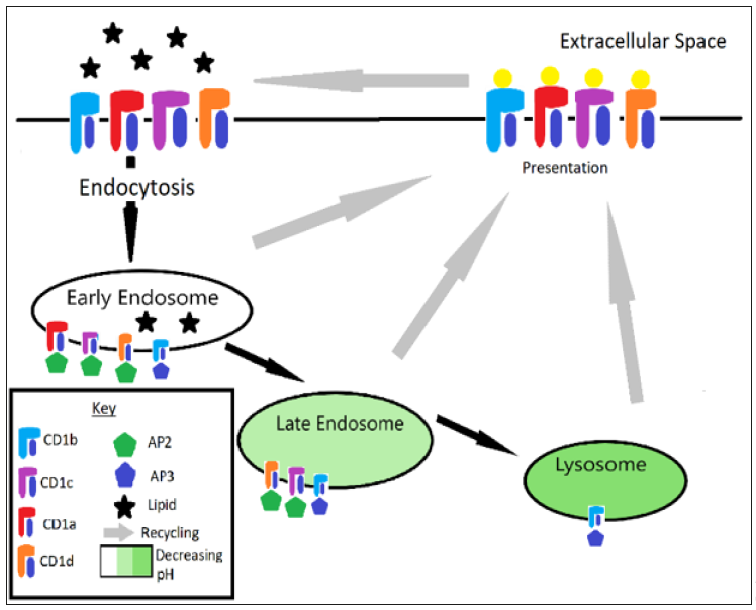
CD1 is utilized to sample the extracellular environment for pathogenic lipids. Additionally, each CD1 isotype seems to have unique targets given that they each localize to different parts of the vesicular system. The system is well described by Chancellor et al.: “Upon assembly in the endoplasmic reticulum, CD1 molecules initially bind polar and non‐polar self‐lipids, believed to function as stabilizing chaperones. Upon correct folding in the endoplasmic reticulum, CD1 complexes travel through the secretory pathway directly to the plasma membrane. Once expressed, they are internalized into lysosomal compartments where they can encounter self and bacterial lipid antigens before recycling back to the plasma membrane. CD1a localizes predominantly in the early endosomes and does not require the low pH conditions for lipid loading. CD1b is primarily located in the late endosomes and lysosomes, where the acidic conditions regulate lipid loading by altering ionic tethers in the F’ portal. CD1c and CD1d recycle to early and late endosomes, as a result of binding adaptor protein 2 (AP‐2) via their cytoplasmic tails” [8]. See Figure 1.
Figure 1: CD1 recycling pathway. Note that CD1a gets recycled after the early endosome, CD1c and CD1d get recycled after the late endosome, and CD1b is recycled after the lysosome.
Much of the literature on CD1 focuses on its structure, CD1b receiving the most attention [3,4,8,23]. “Similarly to MHC class I, CD1 molecules possess two deep pockets: A′ and F′. CD1b has two additional pockets, C′ and T′ that allow the binding of lipids with larger hydrophobic chains. CD1b has the larger binding site, composed of four pockets, three of which are interconnected to form a large A′T′F′ super channel. This characteristic confers CD1b the unique ability to bind long-chain mycolyl lipids” [5]. The size of the channel, while is some ways a blessing, presents a challenge for researchers, “the natural antigens for CD1b range in length from C30 to C80, and so include antigens that would not fully occupy the groove, which is estimated to hold C72–76 lipids, as well as those, like C80 mycolates, which apparently exceed the groove volume Studies of long-chain mycolates bound to CD1b directly ruled out lipid trimming, suggesting that they bind in an intact form” [3].
The literature reports that natural spacer and scaffolding lipids which have been found along with natural antigens of CD1b in elution experiments account for this inconsistency in size [3,4,8]. One lipid implicated as having this function is deoxyceramide [3,22,23]. This suggests that different sized lipids are loaded into and removed from CD1b so that the desired antigen can fit [24]. Additionally, portals in the side of CD1b may account for the binding of lipids larger than the cleft which allow for excess chain length to hang outside of the molecule [3,25]. It has been suggested that these spacer lipids along with CD1e (see above), may play a regulatory role in the binding of CD1b to its ligands, pH may also play a regulatory role as it “induces relaxation of the CD1 structure, promoting a more dynamic binding and dissociation of lipids” [5].
Currently, there are two major questions with regards to CD1 complex binding to TCR: how does it occur and with what specificity?
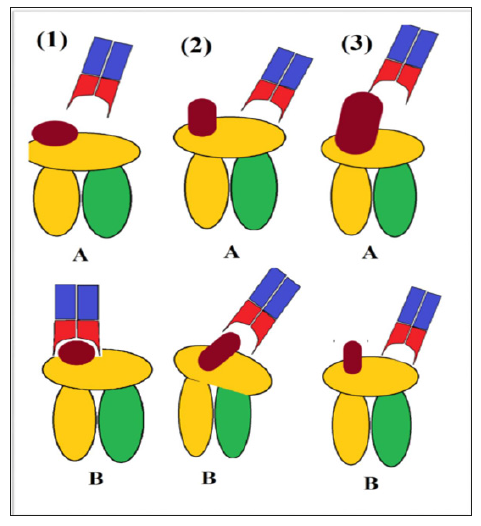
There are currently three models describing TCR recognition of CD1: head group recognition model, conformational remodeling, and absence of interference model [7,26]. In the head group model, the TCR recognizes the head of the lipid in a “tweezer-like mechanism” [8]. Moody et al remarked that this model “emphasized direct and extensive contact of the TCR with antigen” and suggested that “that the TCR can specifically bind to and discern the structure of carbohydrate and other non-peptidic head groups, expanding prior views that TCRs react solely to peptides” [7]. The conformational remodeling and the absence of interference models “both predict that the TCR contacts solely, or mostly CD1 and not lipid”; however, the former also emphasized the dependence of TCR on shape modulation of CD1 for effective binding, whereas the latter merely suggested that TCR naturally binds to CD1 and the ligand merely needs to not block the surface of the molecule [27]. Withing the absence of interference model, two mechanisms have been identified by which lipids could bind CD1a yet fail to interfere with the TCR-CD1a contact: lateral escape, which often shifts the TCR footprint, and seating below the plane of TCR contact, which may still result in a central TCR footprint [7]. See Figure 2.
In addition to contact that the ligand has with the TCR, contacts between the TCR and CD1 may impact binding as well. Chancellor et al. explains that “until recently, a role for the distal or proximal functional groups of the meromycolate tails of mycolic acids as antigenic determinants were not well understood. Van Rhijn et al. showed definitively, a direct impact of distal meromycolate functional group alteration on TCR recognition” [8,12]. Chancellor et al. also notes that “meromycolate chain dynamics within the CD1b groove appear to be directly linked to the activity of TCR”, [28,29] a phenomenon which may be explained by hydrophobic moieties in the crevices of CD1b which ultimately “restricts the head group from adopting positions that facilitate TCR binding.” [8] Van Rhijn et al. similarly reported that changes in mycolic acid (MA) tails resulted in differential TCR binding [12].
Knowledge of the CD1b-restricted T cell repertoire has been slowly growing as the involvement of CD1b in various immunological functions are being established [30]. The role of CD1b in mycobacterial infection was reflected in 2014 by Van Rhijn et al. where they proved that germline-encoded mycolyl-reactive (GEM) T cell TCRs bind with high affinity to glucose monomycolate (GMM), a “mycobacterial glycolipid antigen is produced in vivo during infection and potently activates CD1b-reactive T cells in humans and cows” [2]. Earlier studies showed that invariant natural killer T (iNKT) cells and mucosal-associated invariant T (MAIT) cells selectively bind CD1d [4], and studies have implicated GEM T cells as selectively binding to CD1b [9]. Reflective of their preferences, iNKT cells, as well as MAIT cells, have been shown to have patrolling functions in the body [2,31]. Evidently CD1d has been found to be present on many cell types, both hematopoietic and non-hematopoietic [32]. GEM T cells, on the other hand, have been found to have antimicrobial properties [2], including stimulating the release of IFN-γ [34] and TNF-α [35], and bind to CD1b which is expressed only on professional antigen presenting cells (APCs) such as dendritic cells [8,33].
Studies in the TCRs of MAIT, iNKT, and GEM T cells have reported that the TCR α-chain of these cells types is highly conserved whereas the β-chain is more biased [2,4,9]. This finding, along with crystal structure elucidation, indicate that “individual TCRs could preferentially contact the CD1 protein on the left or instead take a central or right-sided approach to mainly contact the lipid ligand” [4]. Van Rhijn et al. explains that this hypothesis would explain “varied predominant reliance on TCR α- or TCR β-chains for antigen recognition [4]. In defining a novel population CD1breactive T cells, LDN5-like T cells, Van Rhijn et al. noted that their more biased chains have a lower affinity binding for the CD1b-GMM complex whereas the more conserved chains of GEM T cells have a higher affinity for the complex [9]. The linkage between higher conservation and avidity has also been documented [2].
CD1b has offered a lens through which to study autoreactivity, an area only studied through MHC previously: “Precise specificity for the peptide, combined with the processes of thymic education and peripheral tolerance, allows MHC-reactive T cells to minimize reactivity toward self-peptide antigens, and in so doing limits autoimmune disease. Although we have a broad understanding of the molecular mechanisms limiting the MHC-mediated response to self-peptides, the basis of avoidance of self-reactivity toward lipidic autoantigens presented by the CD1 family is less clear.” [13] As reported by Van Rhijn et al., CD1b binds tightly to rare phospholipids, including phosphatidylglycerol (PG), phosphatidic acid (PA), and phosphatidylethanolamine (PE), although affinity is skewed in favor of PG [10].
PG is found in high concentration in the mitochondria [37] as well as on bacteria [38]. While this does technically make CD1b autoreactive, PG is rarely found on the cell membrane [36] and there is therefore little concern of autoimmunity and the benefit of its antibacterial quality remains [10]. Additionally, CD1 group 1 expression is regulated by TLRs, as noted by Chancellor et al.: “Bacterial lipids and products trigger group 1 CD1 expression on dendritic cells through Toll‐like receptor 2 ligation” [8]. However, the autoreactivity is not merely avoided but also use to the immune system’s advantage. Van Rhijn et al. notes that PG is released when the cell is under mitochondrial stress [10]. This may explain why PG accumulates in tumors, as noted by Chancellor et al. [28] which would implicate CD1b as having a role in detecting cancerous cells [8,28].
Shahine et al. Published a detailed model to explain how TCRs recognize autoreactive phospholipids, specifically PG, which is presented by CD1b [13]. The results showed that TCR does not bind to PG independently but binds strongly to the CD1b- PG complex which readily forms. This explanation is compelling, especially in light of the manifold increase in CD1b-PG complex formation when loaded in vitro. However, the authors did not show any attempt to quantitatively correlate endogenous cytosolic PG availability and the frequency of CD1b-PG complex formation in vivo, which is possible and would further support the hypothesis. Nonetheless, SPR steady-state affinity experiments further displayed that PG90 TCR selectively bind PG more over other related phospholipids.
While the head-group recognition model has been accepted for CD1b, the confirmed central footprint of PG90 TCR by Shahine et al. seems unusual given the unequal degree of conservation in the α- and β-chains of CD1b-reactive TCRs, which could have resulted into a right- or left-sided footprint. This can be explained by the high affinity and specificity of the interaction which would block an off-center binding. As proven using structural biology, the PG90 TCR contains a cationic cup which totally envelopes the head group of PG and also interacts with CD1b. Such an overwhelming number of recognition points would neither allow for recognizable dominance of one chain over another in the recognition nor result a recognizably off-center footprint.
Furthermore, it can be emphasizing that one of the gaps in our understanding of the CD1 system is the lack of “understanding of how T cells could discriminate among classes of cellular phospholipids or how abundant self-phospholipids could avoid generating T cell autoimmunity.” It has been determined that, like other TCR interactions, the specificity is determined by specificity in the complementary-determining regions (CDRs) of the TCR, specifically the most central ones – CDR3α and CDR3β, as well as key CD1b residues in the α-helices defining the lip of the cleft [8-13]. Specific moieties and structural shifts, both of the CDRs themselves as well and shifts of the head group induced by TCR, all contribute to the affinity and specificity of the interaction. More concretely, the interaction favors the rare phospholipid PG, whereas more common phospholipids are incompatible, either sterically (as in the case of PC and PI), electrochemically (as in the case of PE and PC) or electrostatically (as in the case of PS). The beauty of this system, similar to PG autoreactivity in its dual-purpose design, as is noted: “Accordingly, it is the architecture of the cationic cup that simultaneously explains the specificity for PG recognition by PG90 and aids in explaining its markedly reduced affinity toward a number of other common cellular phospholipids” [13].
It is therefore evident that there exists a mechanism for autoreactive T cells to be kept alive given certain advantages and precautions. In the case of CD1b, the advantages are mycobacterial pathogen antigen presentation and detection of cells under high metabolic stress. The precautions taken are the tight sequestering of endogenous PG to the mitochondria and the regulation by TLR in activation. In light of the extremely high specificity of the binding and interaction of CD1b-PG and its TCR, another avenue of precaution may be to alter the form of the autoreactive antigen before allowing it to interact with TCRs, similar to the differential binding of lipids based on their MA tails, as noted earlier [28]. (Figure 2).
Figure 2: TCR recognition of CD1. 1) Head group recognition model: A – TCR approaches CD1, B – successfully binds tightly to the lipid. 2) Conformational remodeling model: A – TCR binds slightly to CD1, B – TCR interaction induces conformational changes which allow the lipid to bind. 3) Absence of interference model: A – unsuccessful binding due to steric hindrance of lipid, B – successful binding to CD1 directly due to lack of lipid hindrance.
The CD1 lipid antigen presentation system presents a wide variety of lipids, despite being non-polymorphic, as indicated by the existence of five different isotypes and their unique presentation and recycling patterns in the vesicular system [40]. While most of the isotypes bind ligands with a 1:1 stoichiometric ratio, CD1b seems to bind scaffolding and spacer lipids along with the antigen in its unusually large cleft, a system which allows lipids both smaller and larger than the cleft to be acquired. While CD1brestricted T cells such as GEM T cells have long since indicated an antimicrobial function for CD1b, PG90 TCRs and their specificity for the CD1b-PG complex indicates an autoreactive function as well. While research on the CD1 system has developed with time, the advent of CD1b tetramers has new horizons in experimentation, catapulting the field forward in recent years. As of late, the detailed binding of a CD1b-restricted TCR, PG90, to its ligand, CD1b-PG has been explained [13]. This discovery provides new insight on the mechanism of binding in the CD1 system as well as the potential autoreactive functions of CD1, as indicated by the non-differential binding of both mammalian and non-mammalian PG when presented by CD1b. Future research will explore the role of CD1b in infection, the possibility of using CD1b to present pathogens in vaccines, and the derivation of therapeutic drugs which intersect with the CD1 pathway. The nuanced binding and “plasticity of CD1 antigen presentation paint a complete picture of the cellular lipid environment to T‐cells” will be central in this research [8].


