Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
EG Todorovska*1, D Fasoula2 and I Ioannides2
Received: December 15, 2018; Published: January 03, 2019
*Corresponding author: Elena G Todorovska, Agro Bio Institute, Bulgaria
DOI: 10.26717/BJSTR.2019.12.002300
Barley is one of the major crops for Bulgaria and Cyprus. Knowledge of genetic diversity in barley germplasm helps to ensure that a broad genetic base of breeding materials is maintained not just for sustaining genetic improvement but also for reducing genetic vulnerability to pest, biotic and abiotic stresses. The changeable climatic conditions require precise selection of new cultivars and advanced breeding lines for high yield, tolerance to multiple stress factors and adaptation to different agro-climatic conditions. Responding to the demands for improvement of cereal crops in Bulgaria and Cyprus, a large scale characterization of native and newly introduced valuable germplasm was initiated by molecular markers. The objective of the present study is to compare and explore the genetic diversity in Bulgarian and Cypriot barley collections by SSR markers.
60 accessions of winter and spring two- and six- row barley representing the present-day germplasm from the collections of Agricultural Institute, Karnobat, Bulgaria and Agricultural Research Institute, Nicosia, Cyprus were selected for molecular analysis.
Twelve genomic- and five EST-SSRs [1] were used in the study. Fragment analysis of all SSRs was carried out on ALF ExpressII sequencer and fragment sizes were determined using the computer program Allele Locator v. 1.03 (Amersham Biosciences).
The genetic similarities for each pairs of lines were calculated from a binary matrix using the Dice similarity index. Cluster analyses were performed by UPGMA method with NTSYS-pc ver. 2.11 package. Gene diversity (GD) was calculated using the formula of [2].
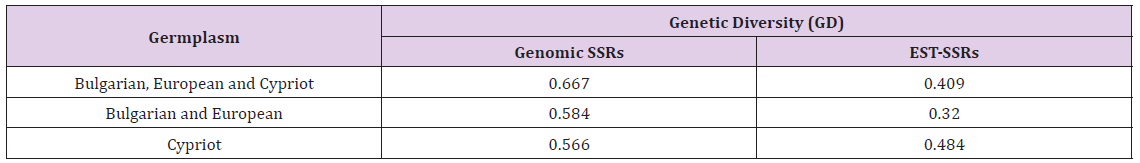
Sixty accessions covering a wide spectrum of genetic diversity of cultivated barley gene pool from different agro-geographical areas (Bulgaria, Central Europe and Cyprus) were characterized at 17 SSR loci. In total, 107 alleles with an average 6.29 alleles/locus were identified with an average GD=0.591. Comparative analysis of gene diversity among germplasm pools from both geographical regions (Table 1) showed higher GD in Cypriot barley in comparison to European barley (0.542 vs. 0.506). Site-of-origin eco-geographic variation of the level of genetic diversity in barley germplasm was observed. European and Cypriot accessions possessing rare alleles at many different loci were detected and could be effectively used for broadening the allelic diversity and the potential for cultivar improvement of barley. The study also clearly showed that the functional diversity revealed by EST-SSRs is higher in Cypriot germplasm collection (GD=0.484) which confirms the role of the variation in the studied transcribed gene regions in adaptiveness to different climatic conditions (Table 2). The constructed dendrograms showed specific grouping of European and Cypriot germplasm pools in two well distinguished clusters (Figure 1). The present study is a base for further analysis of the genomics regions involved in adaptiveness of barley to different agro-climatic conditions and improvement of breeding programmers in Bulgaria and Cyprus by reducing the vulnerability of climate changes and ensuring food security.
Table 1: Description of 17 microsatellite loci, number of alleles and genetic diversity (GD) across 60 Bulgarian, European and Cypriot barley.
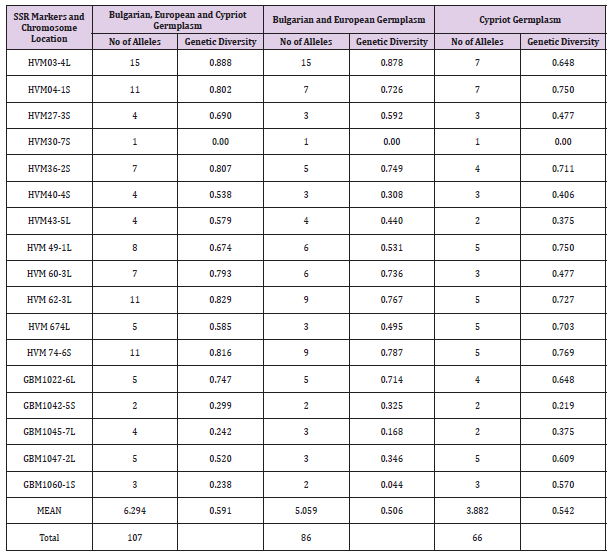
Table 2: Comparison of the genetic diversity in germplasm pools from different agro-geographical regions estimated by genomic and EST-SSRs.