Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Meysam Maghsoodlou1, 2, Mehdi Rajabnia3, Abolfazl Davoodabadi2, Soraya Khafri4, Parisa Sabbagh3, Hamed Ghasemtabar5, Omid Mohammadi2 and Amirmorteza Ebrahimzadeh Namvar2,3*
Received: December 11, 2019; Published: January 02, 2019
*Corresponding author: Amirmorteza Ebrahimzadeh Namvar, Department of Microbiology, Faculty of Medicine, Babol, IR Iran
DOI: 10.26717/BJSTR.2019.12.002282
Introduction: Staphylococcus Epidermidis is known as the most predominant member of coagulase negative staphylococci which can cause nosocomial infections especially in ICU and NICU wards.
Aim: The aim of this study was to investigate the molecular characterization of S. epidermidis strains obtained from the Rouhani Hospital in Babol, Iran.
Materials and Methods: In this descriptive study, a total of 60 S. epidermidis strains were collected. Thereafter, the antimicrobial susceptibility testing, the prevalence of mecA and icaD gene was evaluated. Finally the molecular pattern of isolates was determined by using RAPD-PCR technique.
Results: A total of 50 clinical strains and 10 environmental isolates were obtained from hospitalized patients from different specimens such as bloodstream, urine, catheter, body fluids and etc. by disc diffusion method the high rates of resistance were belonged to oxacillin (70.5%) and ciprofloxacin (63.9%). The prevalence of mecA and icaD genes was reported 85% and 41.6% respectively. 24 RAPD-Type was identified by using RAPD-PCR method which indicates the high genotypic diversity in the S. epidermidis isolates.
Conclusion: The high risk of transmission of infection between the wards and also the Rouhani Hospital staff should be taken seriously.
Keywords: Staphylococcus Epidermidis; Molecular Typing; ICU; Nosocomial Infections
Staphylococci are known as a gram-positive, non-motile and non-spore forming cocci which are divided into two mains coagulase-negative and coagulase-positive staphylococci [1]. Staphylococcus Epidermidis with the highest prevalence rate is recognized as the most significant member of the family of coagulase-negative staphylococci and plays an important role in nosocomial infections [2]. S. epidermidis is considered as the main contributor in medical equipment infections. In recent decades, due to the clinical importance and emergence of methicillin-resistant S. epidermidis strains, this organism has become a challenge in the treatment of patients [3]. Several virulence factors and resistance genes were identified in which the mecA and ica gene are the important one [4]. Many strains of S. epidermidis produce an N acetylglucosamine (PNAG) homopolymer, called PIA which is associated in biofilm formation [5].
PIA is the main element of the extracellular matrix that is produced by the ica gene. The ica gene products include icaA, icaD, icaB and icaC [2]. Identification of these virulence factors is essential in control and prevention of outbreak infections. One of the most effective techniques in this issue is using molecular typing methods. Due to S. epidermidis related infections various typing methods were described such as analysis of plasmid and Restriction enzymes, DNA hybridization, RAPD-PCR, SCCmec typing, PFGE and MLST [2]. As mentioned methods Random Amplification of Polymorphic DNA (RAPD) is a type of modified PCR method in which short-term random sequences are able to attach in different locations of the genome and can produce a range of amplified PCR products [6,7]. The aim of this study was to investigate the molecular characterization of S. epidermidis strains obtained from the Rouhani Hospital in Babol, Iran.
A total of 50 S. epidermidis strains were collected from bloodstream, urine, catheter, shunt, body fluids and wound infections from different wards including open heart surgery, orthopedics, neurology, infectious disease, ICU and NICU of Rouhani Hospital. In addition a total of 10 S. epidermidis strains were obtained from hospital staff for RAPD-PCR test. The isolates were referred to the laboratory of the Babol University of Medical Sciences for confirmation by microbiological test such as gram staining, catalase, coagulase, mannitol fermentation on mannitol salt agar media and sensitivity to polymyxin B and finally stored at -20 °C.
Antibiogram test was performed by disk diffusion method according to CLSI 2016 standards. In this test, Muller Hinton Agar (Merck - Germany) and antibiotic disks included Cefoxitin (30 μg), Oxacillin (1 μg), Gentamicin (10 μg), Erythromycin (15 μg), Clindamycin (2 μg), Ciprofloxacin (5 μg), Fusidic acid (10 μg), Levofloxacin (30 μg), Trimethoprim/Sulfamethoxazole (1.25/23.73 μg), Vancomycin (30 μg), and Linezolid (30 μg) (ROSCO - Denmark) was used.
In order to perform PCR, the MasterMix was prepared in a final volume of 25μl, including: 12.5μl Super MasterMix 2X, 1μl of each primer (Forward & Reverse), 3μl DNA template and 7.5μl Sterile Deionized Water. PCR cycles was perform with initial denaturation temperature of 94°C for 5 minutes, then 30 cycles with a denaturation temperature of 94°C for 30 seconds, annealing temperature of 55°C for 30 seconds, and an extension temperature of 72°C for 40 seconds and a final extension cycle was performed at a temperature of 72°C for 5 minutes. The specific primers are shown in (Table 1).
The DNA genomic was extracted by commercial kit (Yekta-Tajhiz, Iran) and RAPD-PCR assay was carried out according to procedure described previously by Burucoa [8,9]. PCR was performed in a final volume of 25μl containing 12.5μl Super MasterMix 2X, 1μl Primer (5’-CCG CAG CCA A-3’), 3μl DNA Template and 8.5μl Sterile Deionized Water [10]. The PCR program was done as 95 °C for 1 min as an initial hot start followed by 94 ºC for 1 min, 30ºC for 1 min, and 72ºC for 2 min for 4 cycles, followed by 36 cycles of 94ºC for 1 min, 36ºC for 1 min, and 72ºC for 2 min and a final extension step of 72ºC for 5 min. Finally, RAPD-PCR profiles were analyzed by using GelQuest software with 80% similarity cut-off point with clustered using the Dice’s coefficient, and a dendrogram generated based on the Unweighted Pair Group Method with Arithmetic mean (UPGMA).
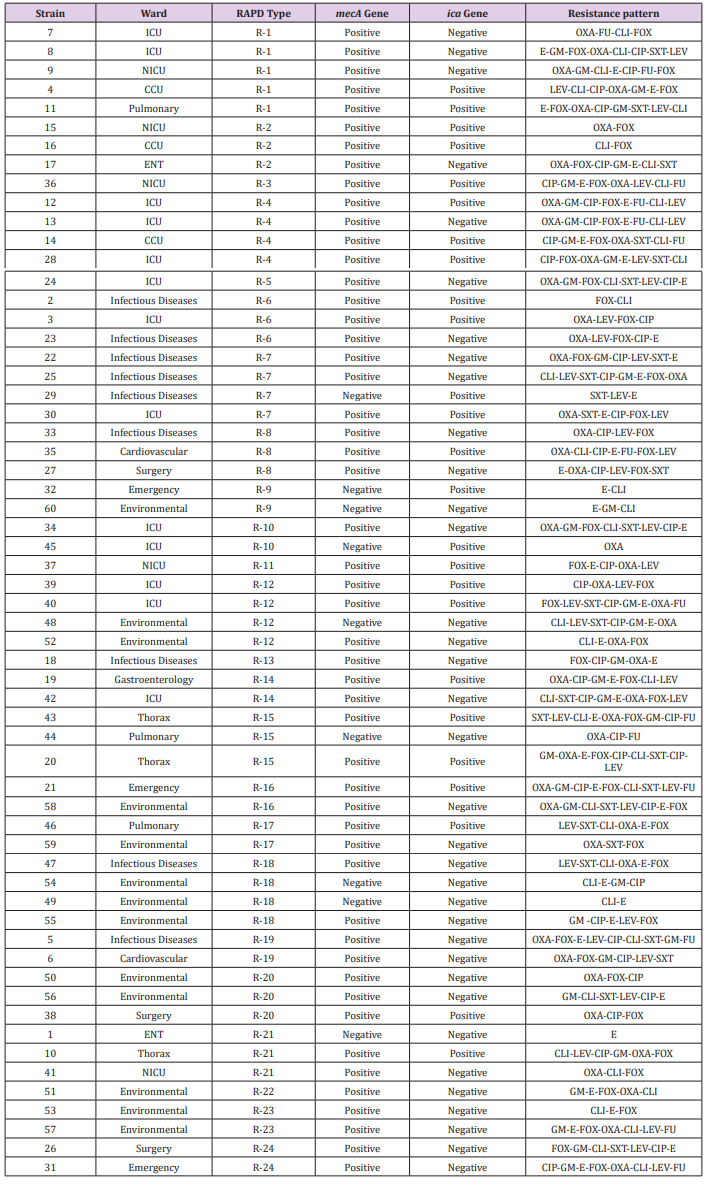
A total of 60 S. epidermidis strains were collected from different wards of Rouhani Hospital. Due to our results the infectious diseases and ICU wards had the highest rate of specimens in comparison to other wards. 53% of isolates were belonged to female gender. By disc diffusion method no resistance was reported to vancomycin and linezolid, while the rate of resistance to oxacillin, gentamicin, cefoxitin, clindamycin, ciprofloxacin and erythromycin were 70.5%, 45.9%, 55.7%, 55.7%, 63.9% and 59% respectively. On the other hand, resistance to trimethoprim/sulfamethoxazole (39.4%), fusidic acid (25.4%) and levofloxacin (41%) was noticeable (Figure 1). The prevalence of mecA and icaD genes was 85% and 41.6% respectively. There were no significant correlation between detected genes and gender (P-value > 0.05). In addition, twenty-four different RAPD-Types were analysis by 80% similarity cut-off point which indicates the high genotypic diversity in our isolates (Figure 2). Table 2 is shown the complete details of all studied isolates.
Table 2:S. epidermidis strains in details including isolation ward, RAPD-Type, mecA and icaD genes and resistance pattern..
According to the results, the presence of mecA and icaD genes among S. epidermidis strains obtained from Rouhani Hospital in Babol was 85% and 41.6%, respectively. The frequency of these genes among similar studies is reported differently. In a study conducted by Najar-Peerayeh et al. The presence of the mecA gene was 92.2% [11]. In the study of Pourmand et al., the frequency of mecA gene was reported 95%, which is higher than of all conducted studies in Iran. Also, resistance to clindamycin and erythromycin in this study was 77% and 79%, respectively which is higher than our study [12]. Moreover, in Pishva et al. research in Al-Zahra Hospital in Isfahan, 75.3% of S. epidermidis strains harbored mecA gene [13]. In a study by Soroush et al., on 80 S. epidermidis strains which was collected from children, 41% had icaD gene. In this study 90% of isolates were found multi-drug resistance, in which the rate of resistance to co-trimoxazole was reported 91.2%. On the other hand, no resistance was observed to linezolid and vancomycin [14].
Moreover, the RAPD-PCR results revealed that RAPD-Type 1 with 5 member followed by RAPD-Type 4, 7, 12 and 18 with four member were the main clusters in current study in comparison to RAPD-Type 3, 5, 11, 13 and 22. These results indicated that the variety of S. epidermidis strains in different wards of the Rouhani Hospital in Babol was high, consequently, due to the wide range of pathogenic factors, S. epidermidis related infections is expected in different wards of the hospital. According to (Table 2), the RAPDPCR method showed that a number of RAPD-Types had the same genotypic profiles and also the same antimicrobial resistance patterns such as RAPD-Type 1, 4, 7, etc. while, the isolation wards were different. There is a significant genetic relationship between the member of some clusters for instance RAPD-Type 4. However, a number of RAPD-Types (9, 12, 16, 17, 18 and 20) illustrated that there was coloration between clinical and environmental isolates. Due to our results, the bacterial infection between various wards and also staff hands and patients is a major concern. Although much progress has been made in molecular typing and more precise methods have been developed, such as PFGE and MLST, but RAPDPCR has maintained its importance as a reliable, cost-effective and user-friendly method. We used RAPD-PCR in this study to find the phylogenetic relationship between S. epidermidis obtained from the Rouhani Hospital in Babol.
This study was funded by the research committee of Babol University of Medical Sciences (Grant no. 3479).


