Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Mohamed A Shreadah*1, Nihad M Abdel Monem2, Galila A Yakout2 and Haiam M Aboul Ela1
Received: Octobet 22, 2018; Published: November 13, 2018
*Corresponding author: Mohamed A Shreadah, Marine biotechnology and natural products(MBNP) Laboratory, Egypt
DOI: 10.26717/BJSTR.2018.10.002025
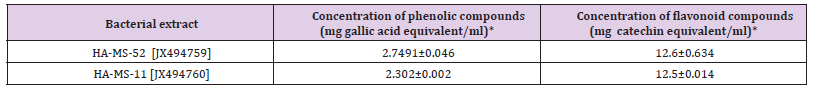
The Red Sea marine sponges have been investigated for isolating novel bacterial strains. The main objective of the present study is to carry out a molecular level identification of the isolated sponge-associated bacteria and examination of their potential to produce biologically active compounds. Sponges were taxonomically identified as Amphimedon Ochracea and as an unidentified species of Amphimedon sp. Taxonomic and phylogenetic analyses on the basis of 16S rRNA gene sequences showed that dominant phylum was Proteobacteria. The results of HPLC analysis revealed that the bacterial exhibited variable patterns of phenolic compounds. Phenolic contents of the examined crude extracts were found to be 2.7491±0.046 and 2.302±0.002 mg gallic equivalent/ml for the HA-MS-52 and HA-MS-11, respectively. Flavonoids contents of the crude extracts of the tested bacterial isolate equivalents of (+catechin) were found to be 12.6±0.634 and 12.5±0.014 for the HA-MS-52 and HA-MS-11 crude extracts. In addition, carotenoid contents tested in the bacterial extracts were Xanthophyll, β-cryptoxanthin and β-carotene.
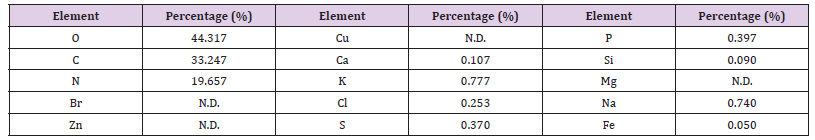
Among these compounds, Xanthophyll significantly showed the highest concentration in both extracts HA-MS-52[JX494759] and HA-MS-11[JX494760] (6.209 & 0.891 μg/ml, respectively); whereas β-carotene exhibited the lowest concentration (0.453 & 0.113 μg/ml, respectively). GC/MS profiling for the ethyl acetate extracts of the studied sponge-associated bacteria revealed the presence of various phytochemical compounds having different biological activities. Results of FTIR spectroscopic studies have revealed the presence of various chemical constituents in the ethyl acetate extract of the HA-MS-52 [JX494759] and HA-MS-11[JX494760] bacterial isolates. Elemental analysis of the ethyl acetate extracts of the HA-MS-52 [JX494759] and HA-MS-11[JX494760] bacterial isolates showed that the bacterial extract indicated the presence of essential elements such as carbon, nitrogen, and oxygen, macro elements such as sodium, potassium and calcium and micro elements such as zinc, The ethyl acetate extract of the HA-MS-52[JX494759] showed un-detectable levels of magnesium, zinc, and bromine.
In the meantime, the ethyl acetate extract of the HA-MS-11[JX494760] showed un-detectable levels of Iron. The results of DPPH test indicated that IC50 values of ethyl acetate extracts had high IC50 values of 1.156 and 2.73 mg/ml for the HA-MS-52 [JX494759] and HA-MS-11[JX494760], respectively. This may be due to the presence of some potent antioxidant compounds which are powerful antioxidants and have good radical scavenging activities. This results were confirmed by the nitric oxide assay (IC50: 5.72±0.002 & 8.96±0.001, respectively). The bacterial extracts showed also potent antioxidant activities using ABTS+ assay with percentage inhibition of IC50 = 0.16 & 8.854 mg/ml, respectively. The high potency of the as strong antioxidant capacity may returns to its high phenolic contents as well as high flavonoids compounds. In the present study highly significant total antioxidant capacity values was also obtained by using phosphomolybdenum assay in the two bacterial extracts suggesting the presence of effective antioxidants in the bacterial extract. The cytotoxicity screening studies revealed the maximum cell growth inhibitory effects of the HA-MS-52[JX494759] bacterial crude extract against the tested cancer cell lines; HepG2; HCT and MCF-7 were found to be weak, 37.5, and 20.7 μg/ml, respectively.
While, the maximum cell growth inhibitory effects of the HA-MS-11[JX494760] bacterial crude extract against the tested cancer cell lines; HepG2; HCT and MCF-7 were found to be 27.5, 21.3, and 7.9 μg/ml, respectively. These results were confirmed by a protein kinase inhibition specialized screening revealing inhibition activities of 87.48± o.62 % and 91.09±0.84 % for the HA-MS-52[JX494759] and HA-MS-11[JX494760] extracts.
Keywords: Sponge Associated Bacteria; Serratia Ficaria; Phylogenetic Analysis; Bioactive Metabolite
Abbreviations: PCR: Polymerase Chain Reaction; GCMS: Gas Chromatography Mass Spectrophotometer; FT-IR: Infrared Analysis; AMD: Age-Related Macular Degeneration; DPPH: 2-Diphenyl-1-Picryl-Hydrazyl Radical
Sponges of the phylum Porifera have been considered as a drug treasure house with respect to great potential regarding their secondary metabolites. Studies have been recently conducted on sponge’s derived compounds to examine its pharmacological properties. Such compounds represent potentially effective agents or lead structures for the treatment of a wide range of human diseases [1-9]. As a consequence of their evolutionary development—mainly regarding their sessile mode of life, their porous and multicelled colonial form, sponges produce a range of secondary metabolites. These compounds are produced in response to the harsh and competitive conditions that occur in the marine environment. These compounds play an important role as allomones and protect them against predators and fouling organisms [10-12]. Based on many studies, sponges have been the largest source of bioactive compounds providing a greater number of novel metabolites than any other marine taxon each year, contributing nearly 30% of all of the natural marine compounds discovered [13]. They harbor a diverse microbial biota representing 35–60% of the total sponge biomass [14].
Natural products derived from marine sponges are diverse in function ranging from anti-inflammatory, antiviral, antitumor, immunosuppressive to antibiotic [15]. Sponges and their associated microbial communities have sparked much interest in recent decades due on the abundant production of chemically diverse metabolites that in nature serve as functional compounds required by the marine sponge host. Production of bioactive natural product by microorganism is a strategy to produce a variety of biologically active natural products in large quantities through the production of microbial culturing. Bioactive natural products from marine bacteria associated with sponges had potency as antibiotics against human microbial pathogen. It also had herbicidal activity and growth promoter activity for plant [16]. The recovery of culturable bacterial strains with different bioactivity profiles shows that the sponge tissues could be rich sources for isolating new strains of bacteria with potential capabilities of producing novel bioactive secondary metabolites [17].
Due to the diverse secondary metabolism these bacteria accumulate a large number of compounds, which may become drug leads for the development of anti bacterials, antivirals, immunosuppressants, antifungals, insecticides, and antitumorals. Red Sea sponges offer a potential for producing novel drugs and prototypes [1-5,7-9]. Several studies have indicated that sponge– microbe interactions are important to produce potential bioactive metabolites in sponge [13]. So far, the bacterial community associated with this key-group in the Red Sea remains unexplored [7,18]. Since it has been reported that more than 15,000 natural compounds with more than 8000 new compounds had been isolated from marine invertebrate where 30% discovered from marine sponges [13], and since most of these bioactive compounds isolated from marine sponges are in fact product of associated microbes [19], the aims of this study are to carry out a molecular level identification of the isolated sponge-associated bacteria and examination of their potential to produce anticancer and antioxidant compounds active against human pathogenic and antibiotic-resistant bacteria.
Sponge samples were collected in May, 2010from four species were collected by SCUBA diving in the Red Sea Figure 1. Three samples were collected from El-Gouna region at depth 1.5 m (GPS: N: 27 22 39.98, E: 33 40 58.95). One sample was collected from the National Institute of Oceanography and Fisheries station, Hurghada at depth 2m (GPS: N: 27 17 07.45, E: 33 46 26.50) Figure 2. The physical, chemical, and biological characteristics of the Red Sea marine environment have been very recently extensively investigated [1-9, 20-35]. Samples were cut from the sponge with a dive knife while wearing latex gloves and individual pieces were transferred to separate plastic sample collection bags, brought to the surface, maintained at ambient seawater temperature and transported to the laboratory in the same day of collection. Sponge specimens were rinsed in sterile seawater, cut into pieces of ca. 1 cm3, and then thoroughly homogenized in a sterile mortar with 10 volumes of sterile seawater. The supernatant was diluted in tenfold series (10−1, 10−2, 10−3) and subsequently plated out on agar plates.
Figure 1: Map of Egypt showing the sampling sites at El-Gouna and Hurghada on the Red Sea coast (A), and the Red Sea (1. Amphimedon Ochraeea and 2. an unidentified species of Amphimedon sp.) sponge.
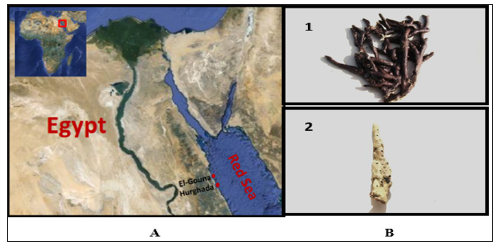
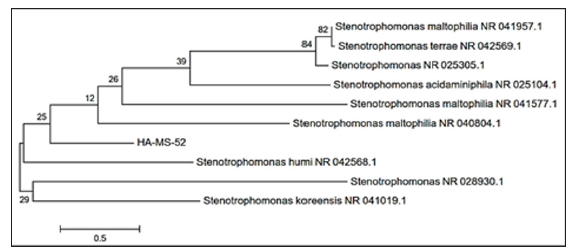
Figure 2: Molecular Phylogenetic anaylsis of the bacterial isolate HA-MS-52 by Maximum Likelihood method. The scale bar indicates 0.5 substitutions per nucleotide position.
The sponge sample was taxonomically identified as Amphimedon Ochracea (Order: Dictyoceratida; Famly: Irciniidae). by Prof. Rob. W. M. van Soest, at the institute for systematic and ecology, the university of Amsterdam, Amsterdam, Netherlands. A voucher fragment of the sponge was incorporated in the collections of the Zoological Museum of Amsterdam, now part of the Netherlands Centre for Biodiversity Naturalis (NCB Naturalis) at Leiden, The Netherlands, under registration number ZMA Por. 22534. While sponge (2) was taxonomically identified by Prof. Michele Kelly at national institute of water and atmospheric research (NIWA) Ltd., Auckland, New Zealand as an un-described species of Amphimedon sp. (Order: Haplosclerida; Famly: Niphatidae). A voucher specimen has been deposited at the Auckland War Memorial Museum. Sponges were transferred to plastic bags containing sea-water and transported to the laboratory. Sponges were rinsed in sterile seawater. Fresh specimen of each sponge was used for isolation of the sponge-associated bacteria. A section of sponge was immediately used to culture the sponge associated bacteria. Another section of sponge was immediately frozen on dry ice and stored at -80 °C. The sponge tissue was then freeze-dried prior to DNA extraction.
Two growth media were prepared to isolate a wide range of the sponge associated bacteria; Marine Agar [36], and ISP medium 2 [37] served as general rich media to grow many heterotrophic marine bacteria. All media were supplemented with 0.2 μm pore size filtered cycloheximide (100 μg/mL), nystatin (25 μg/mL) and nalidixic acid (25 μg/mL). Cycloheximide and nystatin inhibit fungal growth, while nalidixic acid inhibits many fast-growing Gram-negative bacteria [38]. All media contained Difco Bacto agar (18 g/L) and were prepared in 1 L artificial sea water (NaCl 234.7 g, MgCl2·6H2O 106.4 g, Na2SO4 39.2 g, CaCl2 11.0 g, NaHCO3 1.92 g, KCl 6.64 g, KBr 0.96 g, H3BO3 0.26 g, SrCl2 0.24 g, NaF 0.03 g and ddH2O to 10.0 L) [39]. The inoculated plates were incu-bated at 28°C for 5 - 10 days. Distinct colony morpho-Types were picked and re-streaked until visually free of contaminants. Isolates were inoculated into liquid media ISP medium 2. The isolates were maintained on plates for short-term storage and long-term strain collections were set up in medium supplemented with 30% glycerol at −80°C [40].
Bacterial isolates were cultured in 500 mL Erlenmeyer flasks containing 200 mL of ISP2 medium. The liquid cultures were grown for 7 - 14 days depending on their growth rate at 30°C while shaking at 150 rpm. Crude extracts were prepared from whole cultures; containing cells and broths by ethyl acetate equal volume/three times, evaporated, lyophilized and dissolved in 1:1 v/v DMSO/H2O and kept at 4°C until use for cytotoxic assay. The MTT (3-(4,5Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay which Measuring the mitochondrial re-ductase enzyme activity of viable cells that Could reduce MTT to formazan, giving a purple color, was used for screening the cytotoxicity of the bacterial extracts [41]. The cytotoxic activity of the microbial crude extracts was determined against three established cancer cell lines; MCF-7, HepG2 and HCT cells. Based on the values of the IC50, the most potent four bacterial extracts which exhibited the lowest IC50 were chosen for further study of their phylogenetic analysis and identification.
Bacterial DNA was extracted using QIAgen DNAesy Blood and Tissue Kit (Cat. # 69506), following manufacturer’s guidelines: Polymerase Chain Reaction (PCR) Amplification of 16S rRNA. Following extraction of genomic DNA, polymerase chain reaction (PCR) was carried out in order to amplify the 16S rRNA gene, the primers used to amplify the 16S rRNA gene were two universal bacterial primers: Forward primer 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and reverse primer 1492r (5’-GTTACCTTGTTACGACTT-3’) designed to target the conserved regions of the 16S rRNA gene [42].
The isolated bacteria were first identified to the species level by PCR amplification of the 16S rRNA gene, BLAST analysis, and comparison with sequences in the Gen Bank nucleotide database. Specifically, the 16S rRNA gene from the strain was amplified using universal primers 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492r (5’-GGTTACCTTGTTACGACTT-3’). The PCR conditions used were the same as those described previously [43]. The PCR products were purified and sequenced by the GATC-Biotech. Company (Tübingen, Germany). The sequences were compared with known sequences in the Gen Bank nucleotide database and the species level was identified as the nearest phylogenetic neighbor with 98% - 100% similarity [44]. Sequence alignment and phylogenetic analysis were performed using the Mega-5 software package [45]. Tree construction was conducted using neighbor joining algorithm (Jukes-Cantor correction) with bootstrap values based on 1000 replications. The 16S rRNA gene sequences the isolates were deposited in Gen Bank under the accession Numbers indicated in brackets: HA-21 [JQ768238]; HA- 68 [JQ751264]; HAMS- 105 [JQ768239] and HAMS- 119 [JQ768240].
Analytical HPLC Analysis
20 μl sample extract was analyzed with a Exclipse XDB C18 (5 μm, 4.6 *150 mm) column using a mobile phase consisting 1% (v/v) formic acid in aqueous solution: acetonitrile: 2-propanol (70:22:8), pH 2.5; flow rate: 0.75 ml/min, temperature: 30oC, UV detection at 320 nm; Agilent technologies 1200 series. Phenolic compound concentrations were calculated from the standard curves of the supplied phenolic standards exist in the data library.
Analysis was conducted using Gas Chromatography-Mass spectrophotmetery; GC-MS (Trace DSQII MS). MS conditions were as follows: Detector mass spectrometer voltage 70eV and its source temperature was 300o C. The injector temperature was 240oC and the split less mode 0.5μL injection. The HP 55% dimethyl-95% dimethyl tertiary-butyl non-polar column was performed with length 30 cm x 0.25 mm, coating thickness film 0.25 μm. The oven was adjusted at 80°C for 1 min and initial time 1.5 min with 40o C which ended by a final temperature of 300o C and 4 min hold time where the total run time was 15 min. The compounds were identified by comparison with the standards, and also matched with the in-built libraries. The components were identified by comparing their retention times with those of authentic samples, as well as by comparing their mass spectra with those of Wiley 275 Library [46]. Quantitative data were obtained by the peak normalization technique using integrated FID response.
Using Perkin-ELMER1430 infrared spectrophotometer, the molecular structure of the extracts was partial identified. Separated spots of each compound were collected by scratching and dissolved in 1ml ethyl acetate and lyophilized then small discs made from the mixture of about 1mg of the lyophilized material and 300mg of pure and dry KBr were subjected to IR-spectroscope. The measurement were carried out at infrared spectra between 400-4000nm.
The bacterial extracts were subjected to the elemental analysis using Scanning Electron Microscope (SEM) with an energy dispersive x-ray spectrophotometer (EDX). This was carried out at the Regional Center for Mycology and Biotechnology, Al-Azhar University, Egypt.
Total phenolic compounds in the fungal extracts were determined by the method of Taga et al., [47]. One hundred microliters Folin-Ciocalteau reagent (50%, v/v) in distilled water and 2 ml sodium carbonate (2%, v/v) in distilled water were added and mixed well with 100 μl of fungal extract. After 2 h incubation at RT, the absorbance of the resulting blue color solution was measured at 750 nm. Different concentrations (0 – 0.9 mg/ml) of standard gallic acid were used and processed similarly as sample. The concentration of total phenolic in bacterial extracts was determined as mg gallic acid equivalent in 1 ml of the extract using the standard curve of the gallic acid.
Total flavonoid content was determined by a colorimetric method of Zhishen et al., [48]. A 0.25 ml of fungal extract or (+)-catechin standard solution was mixed with 1.25 ml of distilled water, followed by the addition of 0.75 ml of 5% sodium nitrite solution. After 6 min, 0.15 ml of 10% aluminum chloride solution was added and the mixture was made up to 2.5 ml with distilled water and well mixed. The absorbance was measured immediately at 510 nm using a spectrophotometer. The results were expressed as mean ml of (+)-catechin equivalents. Bioactivity screening of the bacterial extracts.
a) Anticancer activity screening.
b) Cytotoxicity against cancer cell lines.
c) Cytotoxicity evaluation using viability assay. For cytotoxicity assay, the cells were seeded in 96-well plate at a cell concentration of 1x104 cells per well in 100μl of growth medium. All experiments were carried out in the triplicate. The cell cytotoxic effect of each tested compound was calculated [49, 50].
.Protein kinase inhibitory effect of the tested bacterial extracts was determined by Universal Tyrosine Kinase Kit; Takara (CAT # MK410) following the manufacturer’s instructions with some modifications. In vitro antioxidant capacity screening.
The extent of inhibition of nitric oxide radical generation in vitro was followed as per the method reported by Green et al., [51].
DPPH radical scavenging assay of the extracts was performed by using modified previously established methodology by Blois [52] and Amarowicz et al., [53]. Assays were performed in flat bottom polystyrene 96 well microtiter plates. To 100 μL of each sample (1-6mg/ml) in EtOH 25 μL DPPH (1mM) in ethanol was added. The resultant mixture was briefly shaken and maintained at room temperature, in the dark for 30 min. At the end of this period the absorbance (A) of the mixture was measured at 490 nm, using Microplate reader spectrophotometer. Scavenging ratio of DPPH assay was calculated as follows:
%scavenging = [(A control - A sample]/ A control)] x 100. ABTS+ Scavenging Effects The antioxidant effect of the leaf extracts was studied using ABTS (2,2’-azino-bis3-ethylbenzthiazoline-6- sulphonic acid) radical cation decolourisation assay according to the method of Shirwaikar et al. [54].
Inhibition (%) = (Control – test ) / Control × 100
All Measurements were expressed as Trolox equivalents. Standard regression line for Trolox was used to calculate TEAC values.
This assay is a spectroscopic method for the quantitative determination of antioxidant capacity, through the formation of phosphomolybdenum complex.
All experiments were conducted at least with 3 replicates and the results were expressed as mean of 3 independent experiments + SEM calculated using Graph pad prism version 6.0.
Culture based techniques and 16S rRNA based molecular technique were employed to characterize the bacterial communities associated with the Amphimedon Ochracea and the unidentified species of Amphimedon Red Sea sponges having promising secondary metabolite profiles. Based on the morphological characteristics and uniqueness, a bacterial pure isolates were selected from the Amphimedon Ochracea and the unidentified species of Amphimedon. The bacterial isolates were given codes prior to identification as (HA-MS -52) and (HA-MS -11), respectively.
DNA base composition and sequencing of the 16S ribosomal DNA (rDNA)
The 16S rRNA genes were amplified by using general bacterial primers. The forward primer (27 f 5’-CCAGAGTTTGATCCTGGCTCAG-3’) and reverse primer (1492 r 5’-TAC CTT GTT ACG ACT T-3’) were used for amplification of 16S rRNA genes the studied bacterial isolate. The amplified 16S rRNA gene was 1.5 kb.
Table 1: Morphological characteristics and code numbers of the isolated bacteria from the sponge samples.
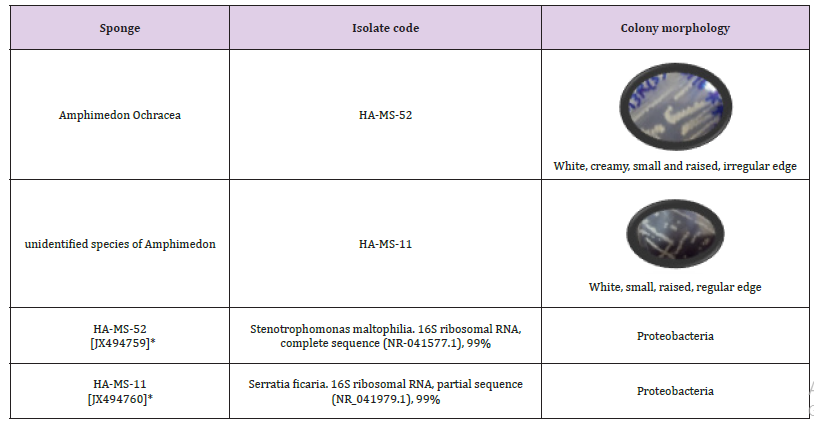
Note: Accession numbers obtained for the bacterial isolates upon submission in the GenBank
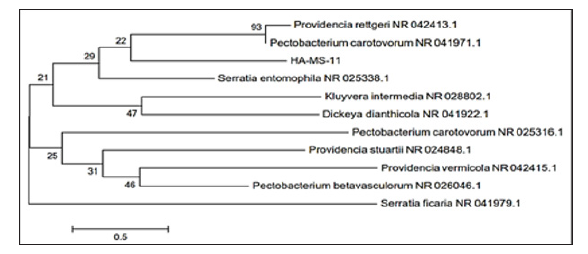
The bacterial isolates were identified using a molecular approach. The results showed that the 16S rRNA genes belonged mainly to the phyla proteobacteria. The sequences were deposited in the Gen Bank database. Accession numbers [JX494759] and [JX494760] corresponded to the strain (HA-MS -52) and HA-MS-11, respectively (Table 1). For the Amphimedon sp. sponge sample, bacterial PCR product was sequenced and phylogeneticaly analyzed (Figures 2 & 3). The present study revealed that the identified bacterial species isolated from Amphimedon Ochracea and unidentified species of Amphimedon was Proteobacteria (especially Gamma- Proteobacteria). The phylum Proteobacteria are always found in different sponges from the same or different geographic location. Proteobacteria have been suggested to have varied effects on sponge hosts such as nitrogen fixation and manipulating host reproduction. It has been reported that Proteobacteria produce low molecular-weight biological active compounds with bioactive properties.
Figure 3: Molecular Phylogenetic anaylsis of the bacterial isolate HA-MS-11 by Maximum Likelihood method. The scale bar indicates 0.5 substitutions per nucleotide position.
Proteobacteria were also found to produce enzymes at high levels for degrading protein and polysaccharides [55]. So far different groups of antimicrobial bacteria including α, β and γ proteobacteria have been isolated from different sponges from geographically different regions [56]. Chemical investigation of the bacterial extract.Ethyl acetate extract of the bacterial isolate was subjected to chemical profiling and characterization using HPLC, GC/MS, FT-IR and elemental analysis techniques.
Phenolic Compounds
The obtained results from the analytical determination of reversed-phase (RP) HPLC against wide range of phytochemical standards revealed that the bacterial exhibited variable patterns of phenolics compounds (Figure 4). The obtained results from the analytical determination of reversed-phase (RP) HPLC against wide range of phytochemical standards revealed that the bacterial exhibited variable patterns of phenolic compounds (Table 2). Chlorogenic acid, Coffeic acid, 3,5-Dicaffeoyl quinic acid, and 4,5-Dicaffeoyl quinic acid were the main phenolic compounds present in the HA-MS-52 [JX494759] crude extract, whereas Chlorogenic acid, Coffeic acid, and 3,5 Dicaffeoyl quinic acid were present in the HA-MS-11 [JX494760] crude extract. Caffeic acid which is a widely distributed phenolic acid prevented the formation of the hydroxy free radicals through the formation of a complex with iron thereby inhibiting the oxidative damage in a dose dependent manner and thereby inhibit the lipid peroxidation. has antiinflammatory, anti-mutagenic, antibacterial and anti-carcinogenic properties, which could be linked to its antioxidant activity.
Note: ND: not detected. All analyses were mean of triplicate measurements.
Figure 4: Main phenolic compounds found in the ten tested bacterial extracts. (A) Chlorogenic acid,(B) Caffeic acid, (C) 3,5- dicaffeoyl quinic acid and (D) 4,5- dicaffeoyl quinic acid.
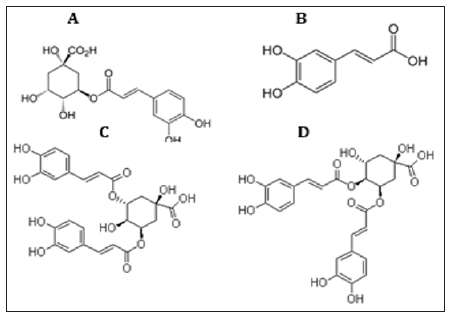
Caffeic acid is able to reduce lipoperoxyl radicals (ROO•)–by donating a hydrogen atom–to its corresponding hydroperoxide, which inhibits the lipid peroxidation chain reaction. In addition to its antioxidant capacity, studies have shown that CA has anti-inflammatory, anti-mutagenic, antibacterial and anti-carcinogenic properties [57]. It also shows immunomodulatory and antiinflammtory activity. Previous studies have shown that caffeic acid inhibits carcinogenesis [58]. It has been stated also that Caffeic acid phenethyl ester suppressed the proliferation of human prostate cancer cells, which strongly suggested the potency of Caffeic acid and its derivatives as anticancer compounds [59]. Interestingly, caffeic acid derivatives such as chlorogenic acid (D-caffeoylquinic acid) which is an ester formed between caffeic acid and L-quinic acid, and it’s isomers of chlorogenic acid include 4-O-caffeoylquinic acid (cryptochlorogenic acid or 4-CQA), 5-O-caffeoylquinic acid (neochlorogenic acid or 5-CQA), 3,4-dicaffeoylquinic acid and 3,5-dicaffeoylquinic acid have also been reported to have strong antiviral activities including HIV, Influenza, Hepatitis B, herpes simplex viruses HSV-1 and anti-inflammatory activity other than their potential antibacterial, anticancer and antioxidant activities [57].
Two of the most powerful structurally related phenolic compounds are Gallic and Tannic acids were also determined by the HPLC analysis for the tested bacterial extracts. Extract HA-MS-52 [JX494759] contained both gallic and tannic acids. In the meantime, extract HA-MS-11 [JX494760] did not contain either gallic or tannic acids (Table 3). Tannic and gallic acids are two commonly phenolic acids that are structurally related. Tannic acid, a naturally occurring plant polyphenol, is composed of a central glucose molecule derivatized at its hydroxyl groups with one or more galloyl residues, whereas gallic acid is a trihydroxybenzoic acid, also known as 3,4,5- trihydroxybenzoic acid. It has been reported that reduced cisplatin induced functional and histological renal damage (Figures 4 & 5). The presence of two flavonoid compounds was tested in the two bacterial extracts; Catechin and Rutin (Figure 6). Catechin and Rutin compounds were below the detectable limit in the extract HA-MS-11 HA-MS-11 [JX494760]. Significantly high Catechin and Rutin values were found in HA-MS-52 [JX494759] extract (15.607 & 2.394 mg/ml, respectively).
Note: ND: not detected. All analyses were mean of triplicate measurements.
The presence of two flavonoid compounds was tested in the two bacterial extracts; Catechin and Rutin (Figure 6). Catechin and Rutin compounds were below the detectable limit in the extract HA-MS-11 HA-MS-11 [JX494760]. Significantly high Catechin and Rutin values were found in HA-MS-52 [JX494759] extract (15.607 & 2.394 mg/ml, respectively).
It can be seen from (Table 4) that the total phenolic and flavonoids values showed high in the ethyl acetate of HA-MS-52 [JX494759] extract compared to HA-MS-11 [JX494760]. Phenolic contents of the examined crude extracts were found to be 2.7491±0.046 and 2.302±0.002 mg gallic equivalent/ml for HA-MS-52 [JX494759] and HA-MS-11 [JX494760], respectively. Flavonoid contents of the crude extracts of the tested bacterial isolates equivalents of (+catechin) were found to be 12.6±0.634 and 12.5±0.014, respectively (Table 5). Carotenoids compounds and Retinol Carotenoids tested in the bacterial extracts were Xanthophyll, β-cryptoxanthin and β-carotene (Figure 5). Xanthophyll, beta-cryptoxanthin, betacarotene were the major carotenoid compounds investigated in the present study. This group of compounds was reported to have strong antioxidant activity and biochemical propertiesinfluencing signaling pathways. Among which, β-carotene was reported to prevent cancers and cardiovascular disorders. Besides, they may protect light-exposed tissues.
Note: ND: not detected. All analyses were mean of triplicate measurements.
Table 5: Total phenolic and flavonoid contents in the bacterial crude extracts expressed as mg gallic acid equivalent / ml.
Note: ND: not detected. All analyses were mean of triplicate measurements./
Ample evidence is available in the literature suggesting betacarotene could possibly lower the risk of stroke, heart diseases, aging, vascular and other metabolic diseases. Carotenoids play a prominent role in protecting bodily cells and thereby act as powerful antioxidants. Moreover, they are proved to be vital for maintenance of proper eyesight and vision. It is now known that beta-carotene is the primary precursor for Vitamin A in humans. Moreover, the positive roles of beta carotene in maintaining a proper vision and its role in other degenerative diseases are now known [60]. There is plenty of evidence now that suggests carotenoids or food products rich in carotenoids are indeed beneficial in the prevention of several types of cancers and malignancies. Certain types of cancers namely, the cancers of prostate gland, urinary bladder and of the colon respond to a natural substance called lycopene. β-Carotene has been used to treat various disorders such as erythropoietic protoporphyria. It has also been used to reduce the risk of breast cancer in women before menopause as well as age-related macular degeneration (AMD).
Interestingly, carotenoids are found in many marine as well as terrestrial sources; however, the positive actions of marine carotenoids are synergistic, and so more powerful, as compared with those from terrestrial origins [60]. Among these compounds, Xanthophyll significantly showed the highest concentration in both extracts HA-MS-52[JX494759] and HA-MS-11[JX494760] (6.209 & 0.891 μg/ml, respectively); whereas β-carotene exhibited the lowest concentration (0.453 & 0.113 μg/ml, respectively) (Table 6) B. GC/MS profiling for the bacterial crude extract: The crude extract of the bacterial isolate was subjected to GC/MS analysis, and they gave peaks as shown in (Figures 6 & 7). Data revealed that, the GC/MS patterns of the bacterial extract by this method 5 bioactive compounds including phenolics, flavonoids, carotenoids and fatty acids. The occurrence of different bioactive compounds suggests a wide range of biological application of the tested extract. The GC-MS analysis of the bacterial ethyl acetate extract showed that sponge is an important source of functional components for the development of new chemotherapeutic agents.
Note: All analyses were mean of triplicate measurements.
Phytochemical investigation of the ethyl acetate extracts of the studied sponge-associated bacteria revealed the presence of various phytochemical such as phenolic compounds, flavonoids. etc. Phytochemical screening serves as the initial step in predicting the types of potential active compounds. The bioactive composition of the bacterial extracts HA-MS-52[JX494759] and HA-MS- 11[JX494760] obtained by GC/MS analysis and their bioactivity profiling are shown in (Tables 7-11).
Note: Number of identified compounds is (7).
Note: Number of identified compounds is (5).
Table 9:Bioactive composition of the crude extracts HA-MS-52[JX494759] and HA-MS-11[JX494760] by GC-MS and their bioactivity profiling.
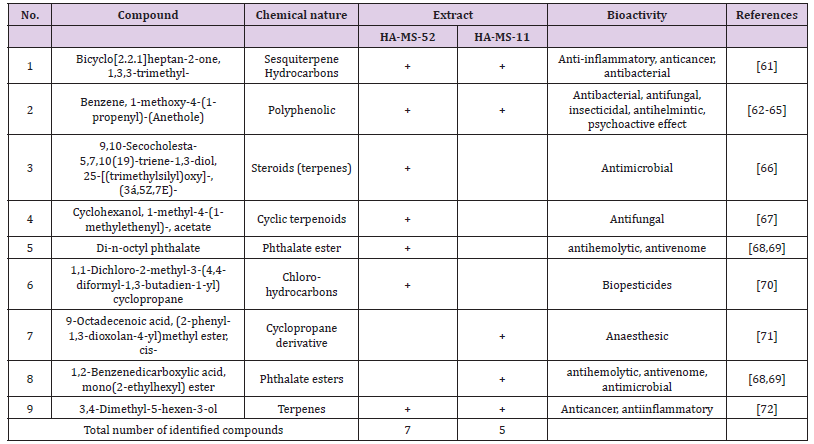
Table 10: Elemental analysis of the ethyl acetate extract of the HA-MS-52[JX494759] bacterial isolate.
Note: ND: not detected. All analyses were mean of triplicate measurements.
Table 11: Elemental analysis of the ethyl acetate extract of the HA-MS-11[JX494760] bacterial isolate.
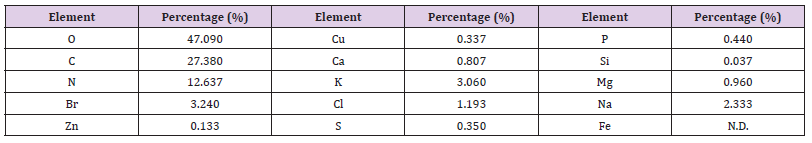
Note: ND: not detected. All analyses were mean of triplicate measurements.
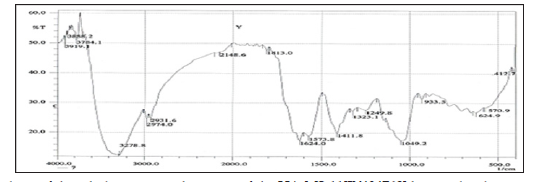
FTIR spectroscopy was useful for the compound identification and when run under IR region in the range of 400-4000 cm-1 there was a variation in the peaks in the tested bacterial extract. FTIR allows detecting the whole range of infrared spectrum simultaneously providing speed and accuracy in measurements of biological specimens. FTIR is one of the most widely used methods to identify the chemical constituents and elucidate the compounds structures, and has been used as requisite method to identify medicines in pharmacopoeia of many countries. Based on the functional group analysis, the bacterial extracts don’t contain any toxic compounds. Furthermore, The IR analysis gave results that suggest the presence of different functional groups. The peak at 3412 and 2926 cm-1 are corresponded to hydroxyl and CH stretching frequency respectively. A band at 1722 cm-1 is corresponded to carbonyl carbon. The peak at 1631 cm-1 to assign C=C. The strong peak at 2927.07 cm-1 assigned to the C-H stretching which means that some alkane compounds existed in the bacterial extract. The stretching assigned to the C–S linkage occurs in the region at 700- 600 Cm-1. The weak absorption band of 601.79, 609.51, 707.88 Cm-1 indicates the presence of physical and chemical (spectral) data concerning FT-IR and Mass analysis obtained in this study were not designed to elucidate the structure of the isolated compounds but to outline and confirm the (Figures 8 & 9) of these compounds.
Elemental analysis of the ethyl acetate extracts of the HA-MS-52 [JX494759] and HA-MS-11[JX494760] bacterial isolates showed that the bacterial extract indicated the presence of essential elements such as carbon, nitrogen, and oxygen, macro elements such as sodium, potassium and calcium and micro elements such as zinc, The ethyl acetate extract of the HA-MS-52[JX494759] showed un-detectable levels of magnesium, zinc, and bromine. In the meantime, the ethyl acetate extract of the HA-MS-11[JX494760] showed un-detectable levels of Iron. The lower concentration of iron (Fe), zinc (Zn), and copper (Cu) is an indication of little or no toxicity of the extracts as heavy metals are known to cause cancer, liver and kidney problems [73]. The elements Mg, Ca, Cu, and Mn are used extensively in chemotherapy and are essential in human and animal health. Deficiency of these trace elements in human subjects can occur under the most practical dietary conditions and in much diseased status. In the recent years, scientists and nationalists have started believing in the therapeutic role of metals in human health. Trace elements play both curative and preventive role in combating diseases.
Calcium is needed in the development of bone and teeth and it regulate heart rhythm, helps in normal blood clotting, maintain proper nerve and muscle functions and lower blood pressure. In the present study, among the various minerals potassium was observed in high level followed by Sodium and phosphorous, this is in agreement with what was reported by [74]. Sodium is the principle extracellular cation and is used for osmoregulation in inter modular fluid of human body [82]. Magnesium is also an active element in several enzyme systems in which thymine pyrophosphate is a cofactor. Oxidative phosphorylation is greatly reduced in the absence of magnesium. It also activates pyruvic acid carboxylase, pyruvic acid oxidase and the condensing enzyme for the reactions in the citric acid cycle [74]. Magnesium is important cofactor for the conversion of blood glucose into energy. Potassium is essential for the transport for nutrients inside the cell. Without potassium, nutrients could not be able to enter into the cell that leads cell death. Silicon is also another important element to prevent the hardening blood veins and arteries.
Chloride works with sodium and potassium carry an electrical charge when dissolved body fluids and to regulate the pH in the body. Chloride is also important for digest the food properly and absorbs many elements that we need to survive. Conclusively, the presence of these elements in the HA-MS-11[JX494760] bacterial isolate marks its usage in therapeutic purposes [75]. Elements such as zinc and manganese are involved in many crucial biological processes and are necessary for the survival of all living organisms because they are important in several enzyme reactions as cofactors, an oxidizing effect, and act as a stringent [76]. The present results revealed considerable amounts of halogens, namely, Br and Cl (Table 13) in the HA-MS-11[JX494760] bacterial extract and only Cl in the HA-MS-52[JX494759] extract. Unlike terrestrial organisms, marine organisms often produce halogenated secondary metabolites, particularly alkaloids [77]. These secondary metabolites not only serve as both initiators and regulators of diverse ecological processes, but they can also be used to treat or probe the causes of human disease. Our data showed that HA-MS- 11[JX494760] bacterial isolate has high bromine contents.
The obtained results were in agreement with [78]. who suggested that marine life produces most of the 4,000 known natural organohalogens, and almost all of the 2,100 natural organobromine compounds are found in marine organisms [77]. The secondary metabolites of living organisms do not exist for the healthcare of human beings. Although the natural products are normally valuable lead compounds, seldom can they be directly used in clinical applications. Halogen containing secondary metabolites exhibit interesting biological effects, such as antibiotic, anticancer and others. This may remark the tested bacterial extract in the current study as promising bioactive materials. Natural products are characterized by their structural diversity, much of which is even unexpected by chemists. Halogenated secondary metabolites, while rare in terrestrial plants, are common in marine organisms due to the abundance of chloride and bromide ions in seawater. Marine biota produces a variety of halogen containing organic compounds that have 1-30 carbon atoms. Organisms that have the ability to form halogenated compounds have been found among various species of algae, bacteria, sponges, mollusks, coelenterates and several marine worms [79].
It has been suggested that new producers of secondary metabolites such as marine bacteria growing on surface or inside marine organisms are expected to produce new halogen containing secondary metabolites [80]. Molecules containing carbon-halogen bonds are produced naturally across all kingdoms of life and constitute a large family of natural products with a broad range of biological activities. The presence of halogen substituents in many bioactive compounds has a profound influence on their molecular properties [81]. HA-MS-11[JX494760] bacterial isolate extract showed high bromine contents in the current study may be promising in their biological and industrial applications. Among halogenated alkaloids, bromoalkaloids were the most widely distributed group of natural compounds. Chemical structures of marine products often differ from terrestrial secondary metabolites in being halogenated with bromine and/or chlorine. The presence of halogen substituents in many natural products profoundly influences their biological activity [77].
On the other hand, (Tables 12 & 13) showed that the two bacterial isolate extracts contained sulfur in considerably high amounts. Sulfur in the form of organic and inorganic compounds is always present in all living organisms and is an important biogenic element. The average content of sulfur is relatively higher in marine organisms. On average natural product contain less nitrogen, sulfur, and halogen atoms but in marine natural product there is relatively higher ratio of sulfur. Sulfur containing phenolic compounds can be reasonably speculated as strong anti-inflammatory and antioxidant agents. Activity of sulfur-containing phenolic antioxidants in vivo is mediated by their effect on redox-sensitive transcription factors. It has been reported that sulfur containing antioxidants are essential in the maintenance of normal well-being of the cell and health [82].
Note: Data are mean ± SD.
Note: Data are mean ± SD.
Antioxidants capacity
Antioxidants are capable of inhibiting the oxidation that produces free radicals. In turn, these radicals can start chain reactions that damage cells. Antioxidants terminate these chain reactions by removing free radicals intermediates, and inhibit other oxidation reactions.
In the present study, the antioxidant properties of the examined crude extracts were demonstrated by DPPH assay. A number of methods are available for the determination of free radical scavenging activity, but the assay employing the stable 2, 2-diphenyl-1-picryl-hydrazyl radical (DPPH) has received the maximum attention owing to its ease of use and its convenience [83]. DPPH scavenging activity in terms of IC50 values as compared to the standard (ascorbic acid) was shown to be relatively high in case of extracts HA-MS-52 and HA-MS-11(1.156 and 2.73 mg/ml, respectively) (Table 13). This may be due to the presence of some potent antioxidant compounds which are powerful antioxidants and have good radical scavenging activities [84]. This finding is also compatible with the total phenolic and flavonoids contents. Nitric Oxide (NO) assay.The antioxidant properties of the examined crude extracts were also examined by using nitric oxide (NO) assay (Table 14). Antioxidants are capable of inhibiting the oxidation that produces free radicals. In turn, these radicals can start chain reactions that damage cells. Antioxidants terminate these chain reactions by removing free radicals intermediates and inhibit other oxidation reactions [85].
Table 14: Total equivalent antioxidant activity and IC50 values of the bacterial extracts using ABTS+ assay.
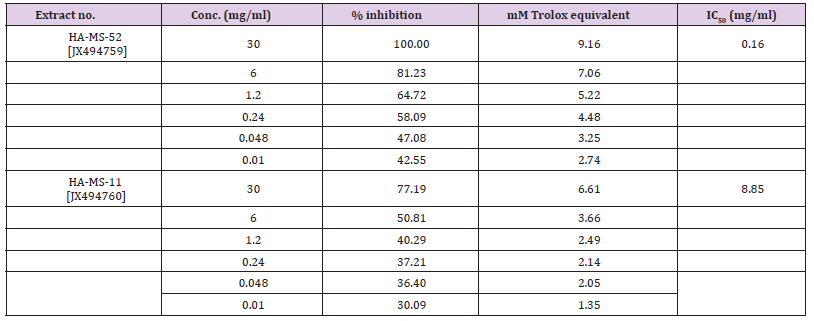
In nitric oxide free radical scavenging assay, highly significant percentage inhibition values were obtained for the studied crude extracts (Table14). IC50 values of the antioxidant activity of bacterial extracts using NO assay were 5.72 and 8.96 % for HA-MS-52 and HA-MS-11 crude extracts, respectively. This may be attributed to the presence of phenolic compounds which have the potential to inhibit NO and peroxynitrite production [86], indicating that the presence of antioxidant molecules in all the tested crude extracts are responsible for their inhibitory effect [85].
The results of the current study cleared out that the ABTS assay gave completely different results for the antioxidant activity measured in the tested two extracts (Table 15). The percentage inhibition of the HA-MS-11 [JX494760] was much higher (IC50 = 8.85 mg/ml) than that of the HA-MS-52 [JX494759] extract (IC50 = 0.16 mg/ml) revealing much higher antioxidant capacity for the HA-MS-11 extract.
The basic principle to assess the antioxidant capacity through phosphomolybdenum assay includes the reduction of Mo (VI) to Mo (V) by the bacterial extract possessing antioxidant compounds. The results of the current study cleared out highly significant TEAC values were obtained in the examined bacterial extracts (Table 14). Addition of various concentrations of the extracts showed that extract HA-MS-52 [JX494760] (47.539±6.99 mM equivalent ascorbic acid at extract concentration 30 mg/ml) was more effective to reduce Mo (VI) to Mo (V), while the lowest effects were shown by HA-MS-11 [JX494759] (31.783±2.87 mM equivalent ascorbic acid at extract concentration 30 mg/ml). The reduction of Mo (VI) to Mo (V) suggested the presence of effective antioxidants in HA-MS-52 [JX494760] bacterial extract.The highly significant antioxidant activities of the tested crude extract HA-MS-52 [JX494760 are mainly due to the presence of biologically active compounds which play an important role in improving human health. The high potency of the tested extract as strong antioxidant capacity may returns to its high phenolic contents [85].
These compounds can decrease the risk of multiple chronic degenerative diseases, for instance gastrointestinal infection, cancer, Alzheimer, cataract and coronary heart disease. Polyphenolic compounds are among the interesting antioxidant compounds isolated from marine resources. It is understood that the intensity of the antioxidant activity of these complex polyphenols is related to the degree of polymerization of the polyphenol. In general, lower degrees of polymerization result in greater antioxidant activities [87].
Cytotoxicity Against Cancer Cell Lines
The utilization of natural products from microorganisms associated with marine sponges which are considered a prolific source of novel cytotoxic natural products with rich chemical structures as the first step in identifying cancer chemotherapeutic drugs. In vitro cytotoxicity assays are a potentially useful tool in the study of toxic compounds of complex mixtures. To explore anticancer drugs from oceanic sources, many researchers have focused their attention on the secondary metabolites of marine microorganisms. By means of cytotoxicity bioassay screening, many novel compounds that are potential candidate anticancer drugs have been isolated from marine micro-organisms for clinical applications since the beginning of this decade [88]. In the present study, the HA-MS- 52[JX494759] and HA-MS-11[JX494760] bacterial crude extracts were subjected to cytotoxicity screening against three established cancer cell lines; HepG2 (hepatocellular carcinoma), HCT (colon carcinoma) and MCF-7 (breast adenocarcinoma). The results of the present study cleared out that the HA-MS-52[JX494759] and HA-MS-11[JX494760] bacterial crude extracts were toxic against those carcinoma cells (Figures 10 & 11). (Tables 14 & 15) revealed that the maximum cell growth inhibitory effects of the HA-MS- 52[JX494759] bacterial crude extract against the tested cancer cell lines; HepG2; HCT and MCF-7 were found to be weak, 37.5, and 20.7 μg/ml, respectively. While, the maximum cell growth inhibitory effects of the HA-MS-11[JX494760] bacterial crude extract against the tested cancer cell lines; HepG2; HCT and MCF-7 were found to be 27.5, 21.3, and 7.9 μg/ml, respectively.
Figure 10: FTIR analysis of the ethyl acetate crude extract of the HA-MS-52[JX494759] bacterial isolate.
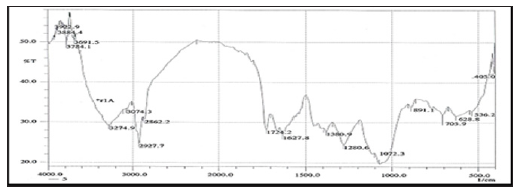
Figure 11: FTIR analysis of the ethyl acetate crude extract of the HA-MS-11[JX494760] bacterial isolate.
Note: Data are mean ± SD.
(Figures 12 & 13) show the cytotoxicity trends of the bacterial extracts against the three cancer cell lines. Interestingly, the high cytotoxic effect of the tested extract may be due to its high sodium content along with the presence of reasonable amounts of caffeic acid. Previous studies showed a potent anticancer effect of sodium caffeate, the sodium salt of caffeic acid, compounds through arresting cells in S-phase [89]. Boron (B) has well established biochemical and nutritional functions. Because of the cytotoxic properties of boron, it became increasingly more frequent in some specific anticancer processes and included in some antineoplastic drugs [90]. It is suggested that more structural elucidation studies to be carried out on specifically on the HA-MS-11[JX494760] crude extract due to its high potency to inhibit all types of the tested cancer cells at low concentrations; low IC50 values. Moreover, data obtained from cytotoxicity test against cancer cell lines are compatible with the high amounts of phenolic, flavonoids, alkaloids and other anticancer natural compounds shown in the tested extract.
Figure 12: Cytotoxic activity of HA-MS-11[JX494760] extract against cancer cell lines; (A): MCF-7, (B): HepG and (C): HCT.
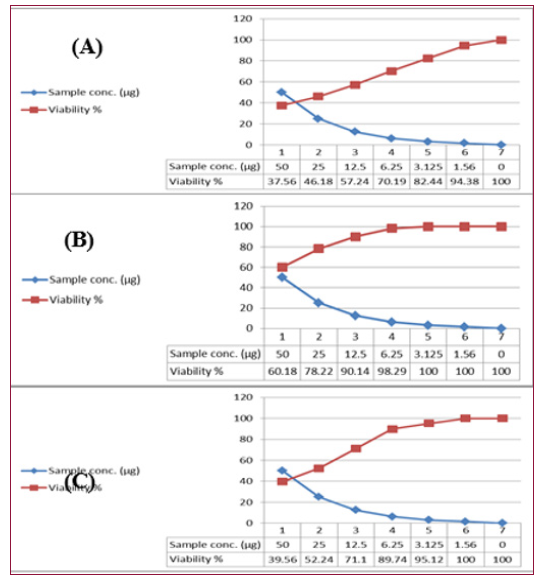
Figure 13:Cytotoxic activity of HA-MS-11[JX494760] extract against cancer cell lines; (A): MCF-7, (B): HepG and (C): HCT.
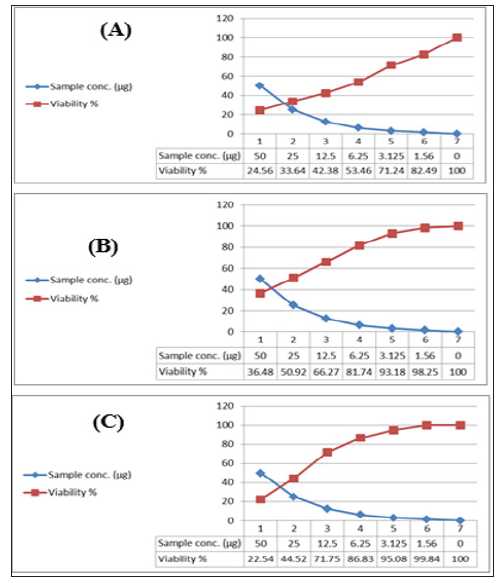
Previous cytotoxicity data were confirmed by a specialized screening of anticancer test; protein kinase inhibition effect. Due to the key roles of kinases in critical signaling pathways, the disregulation of kinase activity has been linked to over 400 diseases including many cancers, autoimmune disorders, inflammation, and diabetes. Therefore, protein kinases are important mediators of cellular communication an d attractive drug targets of many diseases [91]. The present study results revealed that the PTK inhibition activities of the HA-MS-52[JX494759] and HA-MS-11[JX494760] extracts were found to be 87.48± o.62 and 91.09±0.84 inhibition % as a mean of three samples ± S.D., respectively. This may be due to the presence of different alkaloids bioactive compounds. Previous studies showed that natural alkaloids were approved as protein kinase inhibitors [92]. It has been stated that marine sponge metabolites had proven to be extremely potent against a range of kinase targets heavily involved in an increasing list of disease mechanisms including cancer, Alzheimer’s disease and atherosclerosis [93].
Furthermore, as our understanding of the mechanism and regulation of various kinases continues to grow, marine spongederived kinase inhibitors are destined to play an expanding role in the treatment of various diseases. This suggests that further exploration and investigation of these marine-derived protein kinase inhibitors are warranted. Molecular biological and genetic methods together with combinatorial biochemistry of these bacteria also represent a promising approach towards the isolation of new halogen biologically active compounds [94]. From the entire above, marine bioactive compounds could potentially develop as functional food, since their biological activities appear to influence the pathogenesis and the clinical course of several inflammatory diseases (Kadam and Prabhasankar, 2010). Consequently, research should move towards further study and the development of marine functional foods in the hope that, in the future, their regular introduction into the human diet could lead to a reduction in the incidence and severity of many disorders [95].
This work shows that bacterial isolate from the sponge Ircinia echinata and the unidentified species of Amphimedon possess bioactive properties. The tested isolates, HA-MS-52 and HA-MS-11 showed antioxidant activity. It displayed potential cytotoxicity against three established cancer cell lines. Thus this investigation highlights the importance of bacteria associated with the sponge Ircinia echinata and the unidentified species of Amphimedon as a valuable resource for the discovery of novel bioactive molecules. The current study highlighted the functional role of the Red Sea sponge associated bacteria as a good source of biologically active compounds having biotechnological and pharmaceutical significance. Future studies related to their biologically metabolite compounds will elucidate the importance of novel and potential strains associated with sponges.
All the authors gratefully acknowledge Dr. Asmaa Nabiel Adam and Mr. Mohamed Adel from the National Institute of Oceanography and Fisheries, Egypt for their help and technical support. The authors also, acknowledge with thanks Prof. Rob. W. M. van Soest, at the institute for systematic and ecology, the university of Amsterdam, Netherlands and Prof. Michele Kelly at national institute of water and atmospheric research (NIWA) Ltd., Auckland, New Zealand for identification of sponge samples.


