Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Anitha Muthusamy and Leena Grace Beslin*
Received: October 03, 2018; Published: October 17, 2018
*Corresponding author: Leena Grace Beslin, Department of Biotechnology, Selvamm Arts & Science College (Autonomous), Tamilnadu, India
DOI: 10.26717/BJSTR.2018.10.001913
Lipases are produced extracellular by the bacterial strain Bacillus subtilis in submerged state fermentation was purified from the cell free culture broth by ammonium sulphate precipitation at 70% saturation followed by purification solvent precipitation by acetone and ethanol. DS- PAGE analysis indicated that the enzymes protein molecular mass was 19.4 kDa. The enzyme activity was maximum growth 72 hours of incubation time, with an activity (0.95U/ml). Lipase activity was maximal at 1% concentration of the substrate. Olive oil produced (32.67) units of lipase was the best induces for lipase production enzyme activity. The optimum PH 8 for the activity of lipase.
Lipase is produced by various microbes, bacteria, fungi, yeast, mammals and plants in large amounts. Lipase have been used extensively is oleo chemical industry and dairy industry [1] and to produce the structural triglycerides [2]. Extracellular lipase produced by microorganisms is being investigated for its potential application in various industrial processes like detergents, oils, fatty acids and diary coupled with enormous therapeutic uses [3]. Recently, lipases find a number of potential applications in detergent industry, oleo chemical industry, paper manufacturing industry, organic chemical processing, nutrition cosmetics, pharmaceuticals and agrochemical [4]. In leather industry, conventional decreasing is carried out by using kerosene, petrol and other solvents or by aqueous emulsification procedures using detergents. There are many industries pollution problems as well as hazards associated with the use of solvents and detergents. Enzymatic decreasing of hides and skins is suggested as viable alternative to compact pollution problems caused by the use of solvents and surfactants [5]. Microbial lipase is available as commercial production, the majority of which are used in detergents, cosmetics production, food technology and chemical industry. Lipases are biocatalysts because they act under mild conditions, are highly stable in organic solvents, show broad substrate specificity and usually high region and or stereo selectivity in catalysis [6,7] reported that the increased production of extracellular lipase in the culture of microorganism's growth in the presence of triglycerides and other lipids. Some important lipase producing bacterial genera includes Bacillus, Pseudomonas and Aconetobacter. Bacterial lipase is mostly extracellular and are produced by submerged fermentation [8-12].
Multi-faceted microbial lipases (tricyglycerol acylhydrolase, E.C.3.1.1.3) have emerged as key enzymes in swiftly growing modern biotechnology lipase are indispensable for the bioconversion lipids (triacylglycerols) from one organism to another and within the organisms. In addition to their biological significance, lipase have tremendous potential in areas such as food technology, bio medical sciences and chemical industry, detergent [13-15]. Lipases are glycoproteins, but some extracellular bacterial lipases are lipoproteins [16] reported that the enzyme production in most of the bacteria is affected by certain polysaccharides. Many different bacterial species produce lipases, which hydrolyze the esters of glycerol with preferably long chain fatty acids. They act at the interface generated by a hydrophobic liquid substrate in a hydrophilic aqueous medium [17].
In the presence study, the bacillus subtilis was used to production of extracellular lipase by submerged state fermentation by using various substrates. The Bacillus subtilis were identified from oil refineries soil sample from vennandur, Namakkal.
Bacteria are omnivorous, saprophytic, or heterotrophic organisms that occupy nearly every conceivable environmental niche. The soil sample was collected from then oil refineries. Sample was collected by using sterile gloves, scalpel and placed in sterile container. Serial dilution was carried out to isolate bacterial from soil. For this 10ml sterile distilled water was taken in a test tube. To this 1gm of finely pulverized air dried soil was added. The tube was vigorously vortexed for 3 minutes to obtain uniform suspension of microorganisms. A series of tubes, labeled as 10-1 ,10-2 to 10-7 were filled with 9ml sterile distilled water. 1ml of diluted sample was transferred to the tube marked 10-1 and vigorously vortexed. The process was repeated up to 10-7. Pour plate method was used to culture bacteria using nutrient agar media. Three sterile Petri plates was taken and labeled as 10-2,10-3 and 10-4. To these 1ml aliquots each from 10-2, 10-3 and 10-4 serial dilution were transferred to respective Petri plates [18-20]. Then approximately 15ml of cooled (45 °C) nutrient agar medium was added to each Petri plate and the inoculums were mixed by gentle rotation of Petri plates. After solidification of medium the plated were incubated for 24 hours at 37 °C.
The bacillus subtilis culture was grown in tributyrin agar medium composition of medium 2.5g of peptone, 3.0g of beef extract, 1.5g of agar and 1ml of tributyrin was dissolved in 100ml of distilled water. The ph. was adjusted with 7.2.
The tributyrin agar medium was prepared on autoclaved for 1210 cat 15 minutes. The medium was poured into sterilized Petri plates and allowed to solidify and kept for contamination check for 24 hours. The obtained culture was inoculated into two labeled plates one each of ph. 7.2. the plates were kept for incubation 48 hours at 37 °C in inverted position.
The various substrate source of olive oil, neem oil, castor oil, and gingelly oil were used to production of lipase enzyme from bacillus subtilis by submerged stae fermentation. The substrates were obtained from local market at perundurai, erode.
The olive oil borth was used in the present study (the broth contains 5g of peptone, 3g of yeast extract, 2g of beef extract and 5ml of lipid substrates olive oil, neem oil, and castor oil, gingelly oil, dissolved in 100ml of distilled water. The ph. was adjusted with 7.2.
Preparation of Inoculum: Single colony of bacillus subtilis was inoculated in 10ml of olive oil borth in 250ml conical flask incubated at 37 °C with agitation on rotator shaker at 240rpm for 24 hours.
Production of Lipase: 5ml of inoculums was inoculated into 100ml of fresh fermentation medium (the borth contains 2g of peptone, 2g of glucose, 0.5g of potassium dihydrogen phosphate, 0.1g of diammonium sulphate, 0.1g of diammonium carbonate, 0.1g of magnesium sulphate and 1ml of olive oil was dissolved in 100ml of distilled water [21-25]. The Ph was adjusted with 7.2) in 500ml of conical flask and incubated at 37 °C for 96 hours in the shaker at 100rpm.
Extraction of Lipase: After the submerged state fermentation, the enzyme was extracted from fermentation medium, 5ml of production medium was withdrawn from the production flask and centrifuged at 10,000rpm for 30 minutes at 40c. The clear supernatant served as a crude enzyme lipase source.
Enzyme Assay: Lipase activity was assayed by titrimetric (olive oil-substrate emulsion) method [26]. The assay mixture was prepared by 1.0ml of substrate emulsion the composition of substrate emulsion contains 70ml of emulsification reagent (17.6g of sodium chloride, 0.4g of potassium dihydrogen phosphate, 5.4ml of glycerol, 10ml of gum Arabic was dissolved in 1000ml of distilled water) and 30ml of olive oil, 0.8ml of 0.2M potassium phosphate and 0.2ml of enzyme aliquot in 100ml of conical flask and incubated at 550c for 30minutes. After incubation 2.0ml of acetone. Ethanol mixture was added to terminate the reaction and titrate the above aliquots with 0.1N NAOH using 1% phenolphthalein as the indicator, and point is the appearance of pale pink colour. One unit of lipase activity is defined as the amount of enzyme required to liberate 1mg of fatty acid/ml/min
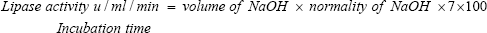
Salt Precipitations: The stability of protein is markedly affected by the ionic strength increase the solubility of the protein also increases this is referred to as "salting in". However, beyond a certain point, the solubility begins to decrease, and this is known as salting out.
Lipase Precipitation by Ammonium Sulphate: Ammonium sulphate used for fractional precipitation of proteins. It was available in highly purified form, has great solubility allowing for significant changes in the ionic strength. The changes in the ammonium sulphate concentration of a solution by adding a solution of known saturation to crude enzyme extract. The 30% salt cut was given to the supernatant was collected from submerged state fermentation. To this 16.6g of ammonium sulphate was added respectively. No precipitation was observed in case of submerged fermentation, the 70% salt cut was added (21.1g of Ammonium Sulphate). Ammonium sulphate was added very slowly with continuous stirring of the solution on a magnetic stirrer in cold conditions. The solution was centrifuged at 10,000rpm for 10 minutes at 4 °C. These pellets were collected and dissolved in 10ml of 50mM Tris Hcl solution.
Dialysis: The precipitate was collected by centrifugation the extract at 10,000 at for 10 minutes at 4 °C the precipitate was dissolved in 10ml of 50mm tris Hcl and subjected to dialysis. About 10cm size of dialysis bag was successively boiled in 100ml of distilled water, 2% sodium bicarbonate and IMM EDTA solution and again 100ml of distilled water for 10min at 100 °C. Then the dialysis bag was cooled to room temperature and kept in refrigerator for 30min. the dissolved enzymes were transferred to dialysis bag of one end. The bag was tightly tied, and dialysis bag was suspended in a beaker containing distilled water with the help of glass rod. This set up was kept in refrigerator overnight.
Solvent Precipitation Using Acetone: The dialyzed enzyme solution was subjected to acetone precipitation at 60% saturation for 30min in the magnetic stirrer. The content of the tubes was subjected to centrifuge at 10,000rpm for 10min, the pellets were dissolved in 5ml of 50MM Tris HCL (PH 8.0) and dialyzed against the same buffer and the enzyme activity was assayed.
Ethanol Precipitation: The dialyzed enzyme acetone precipitation solution was subjected to ethanol precipitation at 705 saturation for 30 minutes in the magnetic stirrer. The content of the tubes was subjected to centrifuge at 10,000rpm 10 minutes, the pellets were dissolved in 5ml of 50MM Tris Hcl buffer (PH-8.0) and the enzyme activity was assayed.
Molecular Mass Determination by SDS - PAGE: PAGE under non - denaturing conditions and SDS - PAGE was carried out as determine by Laemmli. For non-denaturing PAGE, the separating gel consisted of 12% polyacrylamide and the stacking gel consisted of 5% polyacrylamide. Sample of 12ml and 8ul of sample buffer with 50m M Tris Hcl,10 % SDS 0.1% bromophenol blue - mercaptoetanol and glycerol were loaded into the well. Molecular mass markers were purchased from Bangalore genei and were run parallel to the samples. Electric current of 50V was supplied using a standard power pack. The gel was stained using Coomassie brilliant blue and then distained using a mixture of Methanol, Glacial acetic acid and distilled water.
Effect of pH on the Lipase Activity: The effect of PH on different inducers on lipase activity was studied. To determine the optimum PH for the activity of lipase, the medium with different PH were inoculated with culture and incubated at 37 °C for 96 hours. PH ranges of 6-9 were used.
Effect of Substances for Lipase Enzyme Activity: The effect of different inducers on lipase activity was studied by growing the culture in peptone medium at 37 °C for 96 hours. Inducers like olive oil, gingelly oil, Castrol oil and neem oil were used (1%).
The bacillus subtilis was produced the lipase enzyme by checked with lipid hydrolysis with tributyrin agar medium. All the plates were formed the clear zone around the colonies of bacillus subtilis. A clear zone around the colonies was observed due to degradation of fat substrates in the medium. The degradation of fats due to secrete the lipase enzyme by bacillus subtilis.
After 4 days incubation of (production medium olive oil broth) with submerged stae fermentation the crude extract was prepared by centrifuge. The supernatant was used as enzyme source. The crude extract was successively precipitated at saturation of 30% and 70% in magnetic stirrer. The precipitate was collected and dissolved in 10ml of 50mM Tris Hc. The diluted precipitate was subjected to dialysis and the dialysed enzyme was purified.
The bacillus subtilis was cultured in olive oil broth of submerged state fermentation by various substrates of the castor oil, Neem oil and Gingelly oil. The present study indicated that the higher lipase production was observed in olive oil used as a source of lipid substrate [27-31]. The lipase enzyme activity was also high (32.67u/ml/min) in olive oil substances, followed by gingelly oil (28u/ml/min), castor oil (25.67u/ml/min) and Neem oil (23.33u/ ml/min) by assay of olive oil emulsion method. The maximum enzyme production was observed by olive oil > gingelly oil > castor oil > and neem oil respectively (Figures 1-3), when compare to other lipase substrates olive oil was containing high content of lipid substrates were subjected to the bacillus subtilis was easily degradation of olive oil. In the present study concluded the high level of lipase enzyme production, enzyme activity and growth of bacillus subtilis was observed in olive oil broth followed by 32.67u/ ml/min in submerged state fermentation (Tables 1-3).
For determining the molecular mass of the lipase SDS- PAGE (7.5-1.5% polyacrylamide gradient) was performed on electrophoretically purified lipase extracted (by ammonium sulphate precipitation) from slices of non-denaturing gel corresponding to the active lipase bands. Enzyme preparation from different sources were run in parallel to a standard known protein mixture (high molecular weight, kit from sigma) silver staining was used to detect protein bands [32-35]. Silver stains are useful for the detection of Nanogram amount of proteins are nucleic acids in acrylamide gels (or) on various membranes. They are more sensitive than organic stains [36-40]. Silver images of protein or nucleic acid patterns are produced by a difference in the oxidation - reduction potential in regions occupied by nucleic acid or proteins compare to the surrounding gel or membrane [41,49]. This redox potential catalysis the reduction of ionic to metallic sliver. A positive (dark) image will be produced if the region occupied by nucleic acid has higher redox potential than the surrounding region. Experiments with nucleic acids and their components have implicated the purines as the active subunits in the silver staining reaction [50-56]. Estimation of molecular weight of lipase by gel filtration revealed 19000 and31000 daltons respectively. These values correspond to data previously published for microbial lipases [57-62].
the present study bacillus subtilis was used to produce the lipase enzyme by submerged state fermentation.


