Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Malin Jaworski, Johanna Haiko and Baharak Saeedi*
Received: October 03, 2018; Published: October 16, 2018
*Corresponding author: Baharak Saeedi, Division of Clinical Microbiology F 72, Karolinska Institutet, Karolinska University Hospital, Huddinge, SE 141 86 Stockholm, Sweden
DOI: 10.26717/BJSTR.2018.10.001897
Rapid identification of bacterial species causing an infection has shown to have positive impact on the clinical outcome. MALDI-TOF MS is a viable and rapid method for identification of bacterial species not only on agar plates but also directly from positive blood culture bottles and can thus shorten the turnaround time significantly. The aim of this study was to evaluate MALDI-TOF MS on liquid enrichment medium as a possible method for rapid identification of bacterial species by comparing it to routine culturing procedures. The broth was harvested when growth was suspected and analyzed using MALDI-TOF MS. An overall sensitivity of 70.4% was achieved when comparing MALDI-TOF MS and routine procedures. In conclusion, the direct identification from the broth showed a substantial degree of correlation to the routine procedures, where identification was performed from the colonies growing on the agar plates.
To correctly diagnose and treat bacterial infections the type of bacteria causing the infection needs to be accurately identified [1,2]. Rapidly identifying the bacterial species causing an infection has shown to positively impact both the clinical outcome as well as the healthcare costs [3]. MALDI-TOF MS has in recent years become a relatively cheap and easy method of identifying bacteria from solid media and is widely used in clinical laboratories [4,5]. The aim of this prospective study was to evaluate direct MALDI-TOF MS on liquid enrichment medium as a possible method for rapid identification of bacterial species. This was done by comparing the results to those yielded by routine procedures: culturing of clinical samples on solid media and subsequent MALDI-TOF MS analysis on single colonies.
Previous studies have shown that MALDI-TOF MS can be used for identification of bacterial species in different liquid media. Balada-Llasat et al. achieved high levels of sensitivity working with monomicrobial mycobacterial isolates in selective liquid media [6]. While analyzing monomicrobial samples, liquid media shows promise to give reliable results, while polymicrobial samples can present a challenge, as reported by Jadhav et al. who studied a Listeria monocytogenes strain grown in different liquid media [7]. Comparable results have been presented by Lallemand et al. [8] who incubated synovial fluid and osteoarticular samples in blood culture bottles [8]. The samples were treated with Sepsityper Kit (Bruker, Germany) before analysis with MALDI-TOF MS. Even here some difficulties were confronted with polymicrobial samples, but identification of monomicrobial samples showed a high level of success. Lin worked with monomicrobial blood cultures and presented a high success rate [9]. Positive blood cultures where processed in two different steps; either just centrifugation, or a combination of centrifugation and treatment of sample with lysis buffer before application of pellet on a MALDI-TOF MS target. Reich et al. [10] similarly worked with blood culture bottles [10]. The blood culture bottles were harvested and the sample centrifuged and extracted using formic acid before analysis of the pellet. Zhou et al. [11] worked with clinical Escherichia coli, Shigella, and Salmonella isolates alone or in combination grown in liquid non-selective enrichment medium, and received reliable spectra corresponding different species even when present in combinations [11]. However, peak analysis was performed manually, i.e., no automatic identification software was used.
Specimen collection. Samples used in this study originated from deep infections (pus and fluids from the abdomen and thorax) as well as tissue samples, drainage tubes and prostheses. Monomicrobial samples (n=81), negative samples (n=108), as well as polymicrobial samples (n=49) were studied. All samples were cultured into a Fastidious Anaerobe Broth (FAB, LabM) and incubated at 35°C at least overnight or until growth was visible, max. 10 days. In addition to clinical samples, 40 clinical isolates representing 30 unique bacterial species were used in this study. 1-2 colonies of each isolate were inoculated to FAB which was incubated at 35°C and thereafter treated similarly to clinical samples. Sample preparation. According to routine procedures, the bacterial colonies growing on the agar plates were subjected to MALDI-TOF MS analysis, and this result was compared with the direct MALDI-TOF MS result. For direct identification by MALDI-TOF MS, 1ml of FAB was harvested and spun down at 21380g for 1min. Pellet was resuspended and washed in 1ml 0.01M PBS-buffer at PH 7.4 (0.80% NaCl, 0.14% Na2HPO4xH2O, 0.02% KCl, 0.02% KH2PO4) and spun down as above. Supernatant was discarded and 2μl of the pellet was transferred to the stainless-steel target (Bruker Daltonics GmbH) and smeared to achieve even distribution. Sample (either pellet for direct detection or colony for routine detection) was allowed to dry before application of 1μl of 70% formic acid (Sigma Aldrich). Each sample was overlaid with 1μl of HCCA matrix solution (2.5mg a-cyano-4-hydroxycinnamic acid [Bruker Daltonics] suspended in 47.5% LC-MS grade H2O [CHROMASOLV™], 50% LC-MS grade pure acetonitrile [CHROMASOLV™], 2.5% pure trifluoracetic acid [Sigma Aldrich]). Each sample was analyzed in duplicate.
Interpretation of analysis. The spectra achieved were automatically analyzed against the Bruker database using the software Bio typer 3.0 (Bruker Daltonics GmbH). Scores were interpreted with lower cut-off rates than manufacturer's recommendations (1.7-3.0) with reference to earlier studies and evaluations [12-15]. Scores ≥1.7 were interpreted as species-level identification and 1.5-1.69 for genus-level identification. Scores <1.5 were interpreted as unreliable identification. Samples resulting in No Peaks Found (NPF) or scores <1.5 were reanalyzed once and the results with higher scores were used. Statistical analysis. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated using the GraphPad Quick Calcs Web site:_https:// www.graphpad.com/quickcalcs/clinTest1/.
A total of 218 samples, both clinical samples (n=178) and clinical isolates (n=40), were analyzed during this study. Within these, a total of 228 bacterial and Candida species were identified during routine analysis, i.e., identification of colonies by MALDI-TOF MS. Of these microorganisms, 77 were identified as gram-negative rods, 33 as gram-positive rods, 99 as gram-positive cocci and 19 as Candida species. All these samples were also analyzed by direct MALDI-TOF MS from FAB, and the results showed 53.3% agreement of gram-negative rods and 59.3% agreement of gram-positive cocci (Table 1). Among gram-positive rods and Candida species, only 27.3% and 10.5% agreement were achieved, respectively. Polymicrobial samples rarely yielded hits for every microorganism present. In just 3/49 cases two unique bacteria were found with hits >1.5, all three with a different amount (2-5) of microbial species present. For polymicrobial samples the probability of detection was 21% less than that of the monomicrobial samples and isolates (Table 2). In no case did a true negative sample yield a positive result but a mismatch was calculated as a type of false positive. Polymicrobial samples yielded 2 false positives, regarded as mismatches, where Staphylococcus sciuri (score 1.60) was identified by direct MALDI-TOF MS instead of Enterococcus Faecium and Candida Albicans which grew on routine culture. Additionally, Enterococcus faecium (score 1.57) was identified by direct MALDI-TOF MS instead of Pseudomonas Aeruginosa and Candida Albicans on routine culture. These false positive results led to marginally lower specificity and PPV (98.4% and 98.2%). Monomicrobial samples and isolates yielded marginally higher rates of sensitivity (70.9%) when including slow growing microorganisms such as Candida Albicans and Propionibacterium Acnes. Sensitivity was markedly increased when excluding these from the calculations (Table 3).
Table 3: Agreement between routine procedure and direct MALDI-TOF MS on FAB, monomicrobial samples and isolates.
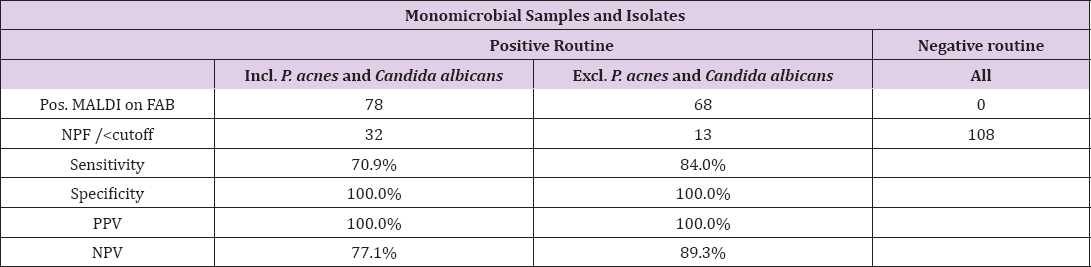
The aim of this prospective study was to examine the possibility to directly with MALDI-TOF MS identify the bacteria growing in a non-selective enrichment broth. It could be useful as a complement to the routine procedures currently in use and could possibly shorten the time for a preliminary result with at least 24 hours in cases where bacterial growth is observed only in the enrichment medium and not in the agar plates. The cutoff of Bruker's MALDI- TOF MS results was lowered from the manufacturer-recommended ≥1.7 to ≥1.5 because this has been done successfully in other studies [13-15]. The risk involved would be to incorrectly identify a bacterial species, but in this study, it only realized twice. Monomicrobial samples and clinical isolates had a high probability of detection (84.0%). Polymicrobial samples yielded polymicrobial results (>1.5) in only 5.9% of cases and no more than 2 unique bacteria were ever found with hits >1.5. This could be because the distribution of microorganisms in an infection is not equal and some grow more quickly and aggressively than others. Additionally, the MALDI-TOF MS instrument is not designed to detect more than one type of organism at a time and if the spectra achieved matched one bacteria, that is what was registered as a hit. In the few cases where correct polymicrobial results were achieved the scores were lower and many were between 1.3-1.49. Sensitivity (70.4%) was low compared to monomicrobial samples and isolates. It was not unexpected that monomicrobial isolates would yield higher scores than polymicrobial samples considering the results achieved by Balada-Llasat, Zhou, Jadhav et al. [6,7,11]. It was, however, interesting to observe that the polymicrobial clinical bacterial samples were suitable for the analysis as well.
Some microbes were more difficult to identify than others, namely Candida species and Propionibacterium acnes. In 15 polymicrobial samples Candida species was present but never identified with direct MALDI-TOF MS. Monomicrobial Candida samples were identified with direct MALDI-TOF MS but never with scores >1.5. Yeasts are known to be more difficult to analyze with MALDI-TOF MS even in single colony form and use of an extraction method to achieve higher scores -which was not done here is common [10]. It is worth to note that Candida did grow in the FAB even though it is not optimal for cultivation of yeasts, but the low sensitivity made it unsuitable to detect with the direct MALDI-TOF MS method. We had 19 clinical samples with growth of Propionibacterium Acnes, and in only 1 of the samples did the score ever reach ≥1.7. In 6 cases, the score achieved was >1.5 and for the rest of them the scores were too low to be considered reliable or no peaks were found at all. The level of growth in the FAB medium for the clinical isolates (n=5) seemed to be significantly higher than that of the clinical samples, which may explain why they performed better with the direct MALDI TOF MS than the clinical samples. It is likely that the results would have been better for the clinical samples if they would have been allowed to incubate longer, but that would have rendered the studied method moot.
Better results might be possible if harvesting more of the FAB, thereby increasing the amount of the bacteria in the pellet. In this study, however, we wanted to treat all the samples identically to make the method more suited to the routines in the clinical laboratory. To highlight the varying results regarding monomicrobial yeast samples and Propionibacterium acnes samples, separate calculations were made. When excluding these samples sensitivity increased from 70.9% to 84.0% and NPV increased from 77.1% to 89.3%. The relatively low NPV (69.7%) calculated for all samples (including polymicrobial samples as well as P. acnes and Candida) is most likely caused by the many NPF results achieved when analyzing polymicrobial samples as well as the slow growing Propionibacterium acnes and Candida species. The FAB medium is not optimal for growth of yeasts.
This theory is supported by the higher NPV (89.3%) calculated when excluding P. acnes and Candida. Possible variables leading to the result of "No peaks found" were many; the applied sample could be smeared too thick, too thin, or not enough or too much material was collected while harvesting the FAB. Patient material could possibly have interfered with the analysis. Even though the pellet was washed it is possible that too much remaining FAB and its proteins could interfere with the analysis as two primary ingredients of the broth are peptone mixture and yeast extract. Additionally, clinical samples naturally vary widely regarding bacterial concentrations, making it more difficult to achieve optimal amount of protein for the MALDI-TOF MS analysis. However, we wanted the method to be as simple as possible and similar to all samples, in order to be used in the clinical routine. Overall, a sensitivity of 70.4% showed that the probability of detection and correlation between methods was high enough for the method to be a viable complement to the routine procedures. Specificity and PPV over 98.0% demonstrates that true negatives would be identified without fail, and the probability to receive a false positive is low. In conclusion, a positive sample might yield a negative result but it was very unlikely for a negative sample to yield a positive result. This could be of clinical significance for acute cases where patients in the intensive care unit can benefit from getting suitable treatment at least 24 hours earlier than not using direct Maldi-TOF from enrichment broth. In many cases correct identification gives valuable information about antibiotic susceptibility and thus potential treatment options [13]. In summary, monomicrobial samples yielded better sensitivity than polymicrobial samples. Overall the calculated sensitivity for all samples was high enough so that the studied method could be considered a reliable complement to routine procedures, and able to give a preliminary result so that appropriate treatment could begin.


