Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Maryam Shafigh1,2 , Ramazan Rajabnia3, Yousef Yahyapour3, Elaheh Ferdosi Shahandashti4, Soraya Khafri5 and Amirmorteza Ebrahimzadeh Namvar2*
Received: August 18, 2018; Published: August 31, 2018
*Corresponding author: Amirmorteza Ebrahimzadeh Namvar, Department of Microbiology, Faculty of Medicine, Babol University of Medical Sciences, Babol, IR Iran
DOI: 10.26717/BJSTR.2018.08.001675
Introduction: Acinetobacter baumannii in the past two decades is known as one of the problematic acquired pathogens in hospitals. The capability of these bacteria in causing multiple diseases is due to emergence of multi-drug resistance strains and compatibility with medical equipments. Aminoglycosides are one of the first-choice drugs used to treat infections caused by A. baumannii strains; however, resistance to aminoglycosides has been developed recently.
Aim: This study aimed to trace prevalence of aminoglycoside modifying enzymes in Acinetobacter baumannii isolates.
Materials and Methods: In this descriptive study, a total of 52 A. baumannii strains isolated from patients of Ayatollah Rouhani and Shahid Beheshti hospitals from Babol. After confirming, an antibiotic susceptibility test was conducted using disc diffusion method. Thereafter the prevalence of aminoglycoside modifying enzymes such as aacC1, aphA6 and aadA1 were analyzed using polymerase chain reaction method.
Results: Amongst 52 isolates, the most resistance was observed in kanamycin, piperacillin, chloramphenicol, rifampicin, cefixime and cefotaxime, also the most sensitivity was belonged for colistin. Moreover, the frequency of aminoglycoside modifying enzymes genes was achieved for aacC1 52(100 %), aphA6 51(98.07%) and aadA1 40(92.72%).
Conclusion: The results of this study showed a significant prevalence of genes encoding aminoglycosides modifying enzymes in Acinetobacter baumannii isolates in the studied region. Therefore, it is important to be concern about the extensive monitoring of antibiotic resistance and the prevention of the emergence of multi drug resistance strains.
Acinetobacter baumannii is a non-spore, non-motile, non-fer- mentable and aerobic gram-negative coccobacilli which belongs to the Moraxellacea family [1]. The most of this genus are opportunistic pathogens in both community and hospital acquired infections in immunocompromised patients, especially in burn and ICU parts after Pseudomonas aeruginosa. These microorganisms have a significant role in various infections including bloodstream, pneumonia associated with ventilation, endocarditis, meningitis, skin, soft tissues, urinary tract and medical implant devices infection. The Acinetobacter species are isolated from numerous sources, such as soil, water, animals and human tissues. Mentioned infections have been effectively treated with traditional antibiotics in the last three decades [2-4]. However, recently the resistance to the most classes of antibiotics such as broad-spectrum penicillin's, cephalosporins, carbapenems, fluoroquinolones, chloramphenicol, tetracyclines and most aminoglycosides has been reported. On the other hand, the emergence of multi-drug resistant strain (MDR) has been a global concern [5,6]. Among various antibiotic, aminoglycosides are considered as one of the critical agents.
Aminoglycosides process is divided in two different steps, first absorption in bacteria which is an important process for bioavailability and second stage is the binding to the ribosome and then inhibition of protein synthesis [7]. Therefore, these groups represent as important antibiotics in the treatment of various Acinetobacter spp. infections, however in recent years, different resistance mechanisms have been occurred [8,9]. The main two mechanisms of resistance to aminoglycosides areas below: first, changing the ribosomal structure such as resistance to streptomycin and second by modifying the enzymatic methods with the presence of genes in bacteria that encodes aminoglycoside-modifying enzymes (AMEs) [10,11]. The AMEs family is classified according to the type of enzyme activity into the three main groups including aminoglycoside, phosphodiesterase transferase (APH), aminoglycoside acetyltransfer- ase (AAC) and aminoglycoside nucleotide transferase (ANT) [10]. Phosphorylation the hydroxyl group of aminoglycosides involves by aminoglycoside phosphotransferase enzyme.
Up till now seven different groups of these enzymes have been identified. The main enzyme group of this family is Aph (3'), which phosphorylate hydroxyl group of the antibiotic in position 34. The frequency of the aphA6 gene in Acinetobacter is widespread and is the cause of resistance to gentamicin, amikacin, kanamycin, paromomycin and neomycin [11]. Moreover, both aadAl and aacCl genes are causing to resistance to streptomycin, spectinomycin and gentamicin respectively [12]. The aim of this study was to evaluate the frequency of aacCl, aadAl & aphA6 genes in Acinetobacter bau- mannii strains isolated from different parts of Babol hospitals.
This descriptive-analytical study was performed on 52 strains of A. baumannii isolated from clinical specimens including tracheal, lung aspiration, sputum, urine culture, bloodstream, tissue infection, body fluid, catheter and etc. from different parts of Ayatollah Rouhani and Shahid Beheshti hospitals. A. baumannii strains were identified by using standard microbiological tests and stored at -200C.
In accordance with Clinical and Laboratory Standards Institute (CLSI document M100-S16) guidelines, antimicrobial susceptibility was performed on the Mueller-Hinton agar plates (Merck, Germany) using the standard disk agar diffusion method for below antimicrobial agents: gentamicin (GM, 10μg), cefotaxime (CTX, 30μg), amikacin (AK, 30μg), tobramycin (TOB, 10μg), ciprofloxacin (CP, 5μg), piperacillin (PIP, 100μg), tetracycline (TE, 30μg), imipenem (IPM, 10μg), meropenem (MEN, 10μg), cefixime (CFM, 30μg), ticar- cillin (TIC, 30μg), colistin (CL, 10μg), chloramphenicol (CHL, 30μg), kanamaycine (K, 30μg), rifampicin (RMP, 5μg) (MAST Diagnostics, Merseyside, UK). After 24 hours, the results were reported as sensitive, semi-sensitive and resistant.
Genomic DNA purification kit (Yektatajhiz, Iran) protocol was performed for DNA extraction. The primers sequences used in this study and PCR program are listed in (Tables 1 & 2). PCR products were subjected to electrophoresis in a 1% agarose gel, and visualized by ultraviolet illumination (Bio-rad, Hercules, USA).
SPSS version 23 and Chi square test was used for statistical analyzing.
Among our study population, 43.2% and 57.7% were male and female respectively. Also, the minimum and maximum age of patients was 34 and 92. On the other hand 48 patients were more than 50 years old (92.3%). The highest number of specimens was collected from post ICU (38.5%) and the lowest was belonged to ENT, Gastroenterology and Organ Transplantation which are shown on (Table 3) in details. In antimicrobial susceptibility test the highest resistance were belonged to piperacillin, kanamaycine, chloramphenicol, rifampicin, cefixime, cefotaxime antibiotics while the colistin was the most sensitive one (Figure 1) By molecular PCR method the prevalence of aacC1, aphA6 and aadA1 genes were determined as: 100%, 98.7% and 76.92%. (Figures 2-4)
Figure 2: PCR of the aacCl gene.
Line 1, 2 and 6: Positive strains with465bp (ampliconsize).
Line 3: 100 bp DNA Ladder.
Line 4: Negative control and Line 5: Positive control.
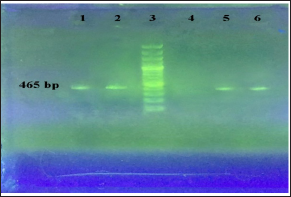
Figure 3: PCR of the aphA6 gene.
Line 1, 2 and 4: Positive strains with 780 bp(ampliconsize)
line 3: 100 bp DNA Ladder
Line 4: Positive control and
Line 5: Negative control.
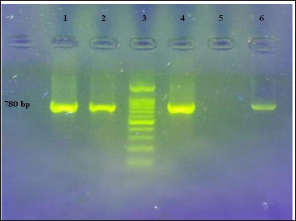
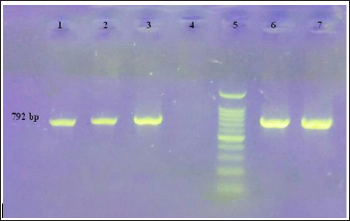
Figure 4: PCR of the aadAl gene
Line 1, 2,3 and 7: Positive strains with 792 bp(ampliconsize)
- Line 4: Negative control
Line 5: 100 bp DNA Ladder and Line 6: Positive control
In the present study, we investigated the prevalence of aacC1, aadA1, and aphA6 aminoglycoside resistance genes and antimicrobial resistance patterns in Acinetobacter baumannii strains isolated from hospitalized patients in Babol hospitals using both phenotypic and PCR methods. Recognition of these genes can be helpful for identification of suitable and effective antibiotics in the treatment of A. baumannii infections for the reason that mentioned microorganism is known as one of the common causes of hospital infections around the world, in accordance to intrinsic and acquired resistance to various antibiotics. Recent studies indicate that the antibiotic resistance pattern of these strains is increasing progressively. In a study conducted by Moniri et al. in Kashan the highest antibiotic resistance to amikacin and tobramycin was 80% and 68.3%, respectively, which is consistent with the recent study [14]. Also, in Hosseini-Jazani et al. the resistance to gentamicin was 70.8%, amikacin 52% and kanamycin 95.8% [15]. In the study of Alaee et al. in Shiraz, all strains showed high resistance to both cefotaxime and kanamycin [16].
In the study of Aliakbarzade et al. in Tabriz, resistance to cefixime, cefotaxime, gentamicin, amikacin, ciprofloxacin and chloramphenicol showed a significant increase while, colistin demonstrated the lowest resistance. Similar to our research, in comparison to colistin all of the strains were resistant to cefixime and also had high resistance to cefotaxime, gentamicin, amikacin, ciprofloxacin and chloramphenicol [13]. In the conducted study by Shahcheraghi et al. in Tehran the resistance to cefixime was the most one in contrast to colistin [17]. It's notable that due to performed researches, the resistance to different antibiotic agents is being increased. However, in Moniri et al. results the prevalence of aacC1, aphA6 and aadA1 were 63.3, 65 and 41.7% [14]. The prevalence of these genes in our study is much higher than of Moniri et al. This difference may indicate the presence of gene cassettes in various A. baumannii strains in our isolates.
In a study by Lee et al. in Korea in 2011, resistance to amikacin and tobramycin was 53% and 67%. The majority of aminoglycoside-modifying enzyme genes that were detected by PCR method were aacC1 (56%), aadB (48%) and aphA6 (71%) [18]. While in Nemec et al. study in 2004, 96% of strains were resistant to at least one of the antibiotics of kanamycin, gentamicin, tobramycin and amikacin. In addition, in 95% of strains the association of the genes of aphA1, aacC1 and aphA6 with resistance to kanamycin, gentamicin, kanamycin and amikacin was reported [9]. Whereas, in our study, 98.7% of strains had (aacC1) and (aphA6) genes, as well as 80.77% of the strains that had all of the three aacC1, aphA6, and aadA1 genes. This variation in the incidence of these genes in numerous regions such as Iran, the Czech Republic and Korea can indicate the spread of different strains that may in accordance with difference distribution of resistance genes among aminoglycosides.
Akers et al. in 2010 evaluated the sensitivity of 107 isolates of Acinetobacter baumannii-calcoaceticus complex to amikacin, gen-tamicin and tobramycin by using disk diffusion method, and reported the sensitivity of 96% of strains to gentamicin and 77.5% tobramycin [19]. In another research conducted by Nigro et al. the resistance pattern of sixty-one multi drug resistant strains of Acinetobacter baumannii was determined and also AME genes like aadB, aacC1, AphA1b, aphA6 and OXA23 beta-lactamase was reported from 2000 until 2010 in 6 Australian hospitals (20). As mentioned above, the rate of MDR strains in our study was considerable, in which 36.53% and 19.23% of strains were resistant to 14 and 13 antimicrobial agents. Furthermore, 3.84% of all strains were resistant to all 15 antibiotics which were used in this study.
Hence, these results indicate that AME-associated genes are common in our Acinetobacter baumannii species. The multiple anti-biotic resistance mechanisms of these bacteria have become a major treatment concern in health care units. In conclusion according to present study the resistance genes among aminoglycoside have been increased and this distribution may be related to geographical area.
The authors declare they have no conflict of interest.
This research was supported by Babol University of Medical Sciences (Grant no. 3485)

