Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Shradha Mukherjee, Ph.D.*1,2
Received: July 07, 2018; Published: July 26, 2018
*Corresponding author: Shradha Mukherjee,Ph.D., USA
DOI: 10.26717/BJSTR.2018.07.001479
Abbrevations: ALT: EHR: Electronic Health Records, SNPs: Single Nucleotide Variations, NGS: Next Generation Sequencing, WGS: Whole Genome Sequencing, WES: Whole Exome Sequencing, GWAS Genome Wide Association Studies, HGMD: Human Gene Mutatuin Database, RNAi: RNA Interference, ZFNs: Zinc Finger Nucleases, TALENs: Transcription Activator Like Effector Nucleases
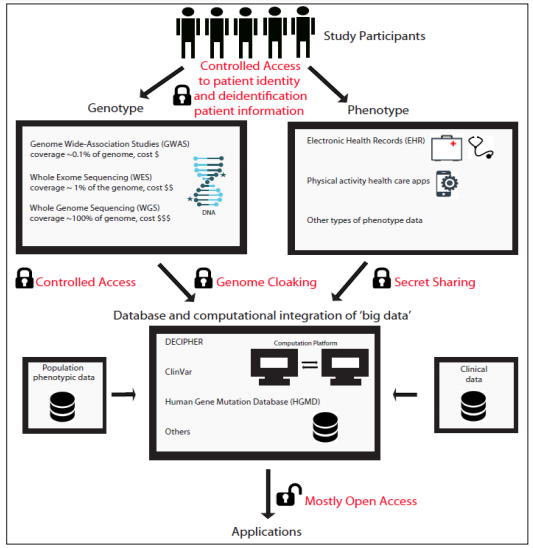
Contemporary studies in human genetics has become a ‘Big Data Science’, due to recent advances in sequencing technologies that has reduced the cost and improved accuracy of sequencing. This high volume ‘Big Data’ consists of genomics data, clinical data, electronic health records (EHR) and physical activity data form personal health apps and presents the unprecedented opportunity to combine them for integrated ‘Big Data’ healthcare analytics and applied knowledge. There is legitimate privacy and security concerns over indiscriminate open source access and use of the biomedical ‘Big Data’, while creation of barriers to data access is feared to hinder research endeavors in human diseases. In this brief commentary, the present status of genetic studies and data security are discussed(Figure 1).
Over the past few decades, analysis of human genome or DNA sequences for identification of disease causing mutations has been one of the greatest focus of genomics research. Identification of disease associated single nucleotide variations (SNPs) in DNA is based on the hypothesis of ‘common disease common variant’. The assumption behind this hypothesis is that a common disease must have a different genetic structure than rare diseases. This assumption was supported by discoveries of susceptibility variants or SNPs with high minor allele frequency, on APOE gene and PPAR gamma gene, for the common diseases, Alzheimer’s disease and Type II diabetes Blacker et al. [1]. These successes combined with the evolution of genomic technology, has ushered an age of genomics with tremendous growth in genotype (DNA sequence and variation) and phenotype (disease manifestation) data. In these GWAS (Genome-Wide Association Studies) projects researchers have identified single nucleotide variations (SNPs) in the human genome associated with diseases.
Identification of genetic variations (genotype) associated with disease (phenotype), also called association studies, involves determination of the DNA sequence. The two technologies available to do this at a genome-wide level, are Array-based technology Distefano et al. [2] and Next-generation sequencing (NGS) technology Goodwin et al. [3]. Array based technology is based on DNA-probes, each ~70 base pairs in size, which recognize specific sites on the genome and emit a measurable signal when there is a match. The Array based technology covers ~500,000 or 1,000,000 (< 0.1% of the genome) different genetic variations or SNPs on the genome. Arrays are available either as an Affymetrix platform with DNA probes printed on a spot of a chip or as an Illumina platform with DNA probes on beads. The human genome is made up of 3.3 billon bases, while the arrays cover only ~500,000 or 1,000,000 sites. Thus, the key to the success of the array based GWAS for identification of disease associated variants, came from careful selection of sites (SNPs or markers) which are known to vary across a population of humans and/or prior studies have shown them to be a disease associated locus. Over 25,000 significant disease-associated genetic loci have been identified so far with the help of Array based GWAS studies Mac Arthur et al. [4].
In NGS bases technology it is not necessary to have prior knowledge of genetic variants in the population, as NGS based Whole Genome Sequencing (WGS) covers all 3.3 billion bases or sites on the genome without any bias of site or marker selection. As the cost of sequencing with NGS continues to decrease this technology is poised to be widely used to identify not only common variants, like the Array based technology, but also rare variants associated with diseases Bennette et al. [5]. Given the cost of WGS, presently the most widely used NGS bases technology is Whole Exome Sequencing (WES), which sequences base by base the entire protein coding region or exons of the genome (~1% of the entire genome). NGS based technology, has revealed that an estimated 100 loss of function variants or 100 non-functional genes occur per human, with around 20 of them being homozygous or completely gene inactivating in each person, and most occur population wide at a frequency of >1% Mac Arthur et al. [6]. Thus, WGS and WEG, have the potential to reveal novel genetic variations associated with diseases.
Since the first human genome draft an immense amount of DNA sequence data and genetic variations that is associated with population differences and diseases have been assembled. Genetic data sharing is integral to enable scientific discovery and interpretation of disease associated genetic variants. Without data sharing resources researchers would have to bear an increased financial burden due to data storage costs and this would hinder ‘big data’ genetics research Sousa et al. [7]. The databases, such as, DECIPHER, Clin Var and Human Gene Mutatuin Database (HGMD) integrate genetic data from GWAS, WES and WEG studies, with phenotypic and clinical data and maintain predominantly open source data storage, and data sharing Stenson et al. [8]. Data from these databases facilitates research for identification of key disease associated variants in DNA sequences from healthy people and patients. This information can then be applied for disease risk assessment in healthy people and for treatment of patients with disease associated genetic variant using corrective gene-editing. For corrective gene-editing, RNA interference (RNAi) based technology, has been successfully applied to correct genetic variants in Wiskott Aldrich syndrome and the application of programmable nucleases-based technologies, such as zinc finger nucleases (ZFNs), transcription activator like effector nucleases (TALENs) and Crispr Cas9 are in the horizon Cox et al. [9].
Identification of genetic variants associated with diseases, holds great promise for risk assessment and targeted therapy. However, the potential of DNA sequencing for identification of genetic variants is clouded by privacy concerns and its potential for discrimination based on health status disclosure Shi et al. [10]. In one of the well-known examples that demonstrates privacy concerns around genetic sequencing, surrounds the release of genome sequence of Dr. James Watson, the co-inventor of DNA structure. Watson requested that the sequence of his DNA around APOE gene, variations in which is associated with Alzheimer’s disease, be kept private and out of the otherwise publicly shared Watson’s Whole Genome Sequence (WGS). Scientists soon pointed out that genetic variations in sequences of genes around the APOE gene, with linkage disequilibrium between one or more polymorphisms and APOE gene, were sufficient to predict the state of APOE gene variants Nyholt et al. [11]. This resulted in removal of an additional chuck of open source Watson DNA sequence from the database. Indeed, protection of genetic data obtained from healthy participants and patients from privacy breaches is the top concern and barrier in enrollment of people for genomic sequencing studies Mc Guire et al. [12]. The success of global genetic sequencing studies for identification of disease associated variants, largely depends on collection of genetic information across multiple samples across the human population. Therefore, it is critical to alleviate privacy concerns of people to increase participation and maximize collection of genetic information across the population.
Controlled Access Method: The most common method used to address the privacy concerns of data sharing is extensive vetting to control and limit the access to DNA sequence information, while summary statistics are shared more freely Dankar et al. [13]. Deidentification of source or individual from whom the sequence originated and instead sharing metadata, such as basic demographics and health condition, further potentiates security Dankar et al. [10]. The same DNA sequence and metadata information that makes genetic sequence-based precision medicine effective, can potentially also be utilized for identification of the individual patient. For example, it has been shown that the birthdate, gender and zip-code available in the metadata can be used to identify from which individual the genetic sequences originated even in deidentified patient data Dankar et al. [14] while likelihood-ratio tests have been shown to predict if an individual’s genome sequence is part of the database from the sequence of a single allele Shringarpure et al. [15]. Deidentification of individuals combined with the metadata on their health condition poses great privacy concerns of exposure from database attacks.
Genome Cloaking Cryptography Method: To secure against deidentification database attacks and to maximize beneficial use of DNA sequence data, a more sophisticated method of genetic data encryption has been developed by computer scientists and mathematicians Jagadeesh et al. [16]. In cryptographic ‘genome cloaking’, genetic variations in each genome is converted into linear series of values and only the genetic variations relevant to the query or research question, such as what the genetic variations are associated with a certain disease are revealed to the user, while the remainder of the genetic sequence is hidden from the user. Therefore, the input (entire genetic sequence) is not revealed to the user and only the output (relevant disease associated genetic sequence differences) is revealed to the user, and prevents any private genetic information being revealed.
Secret Sharing Cryptography Method: Recently, another cryptographic method was developed called ‘secret sharing’. In this technique, the sensitive genomic data is shared among multiple servers in such a way that stored data in one server is not complete without data on the other server Cho et al. [17]. For example, if genetic information at a particular SNP or DNA site is stored not as a single value, but as a subtraction value equal to (x-r), where ‘r’ is a randomly assigned number stored in Server 1, while ‘x’ stored in Server 2 is a number such that the difference of ‘x’ with the value of ‘r’, gives the genetic information at that genomic location. Therefore,for data protection only the Server 1, which houses ‘r’ values need to be secured, while Server 2 can be made more open access. This method to secure only part of the information, is much more cost effective that other cryptographic methods, such as ‘genome cloaking’, which require securely storing the entire information(Figure 1)
Figure 1: Overview of types of biomedical ‘big data’ sources, storage, integration, application and security. Study participants (control and patients) are the sources of genotype (GWAS, WES or WGS) and phenotype information (EHR and/or health apps data). This data is then stored and compared with preexisting knowledge bases (population phenotypic and population clinical data) to derive actionable inferences that can be applied for risk assessment and provide treatment solutions for the study participants.
Lower costs of computing and sequencing technologies has made genomic data easier to collect, store and process. Integration of genomic data with other ‘Big Data’ such as, clinical data, electronic health records (EHR) and physical activity data form personal health apps, is making genomics data more powerful and adept at risk assessment and identification of disease associated variants. There is extensive support for research focused on the utilization of genetic sequence data integrated with other ‘Big Data’ for personalized medicine, risk assessment and forensic investigation. Albeit, there has been a parallel growth in privacy concerns over genomic data sharing and utilization. In this brief commentary, an overview of the state of art of genomic data collection, storage, use and advances in computational methods for data protection have been discussed. Controlled access and data encryption methods have come a long way and hold the key to prevent inappropriate use and privacy breaches of genomic data, while it will still allow the biomedical field to design breakthrough therapies from the integrated ‘Big Data’ analysis of genomics and other healthcare ‘Big Data’.


