Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Zhao Haiping1, Chen Guangxin2, Li Haitao*1 and Li Chunyi*2,3
Received: June 15, 2018; Published: June 25, 2018
*Corresponding author: Li Haitao, Nanjing University of Chinese Medicine, Changchun Sci-Tech University, Nanjing, 210029, China
Li Chunyi, Nanjing University of Chinese Medicine, Changchun Sci-Tech University, Nanjing, 210029, China
DOI: 10.26717/BJSTR.2018.06.001294
Intracellular sodium ion functions as an initiating signal for organ regeneration in lower vertebrates, such as amphibians, Intracellular sodium ion concentration ([Na+]) was regulated by voltage-gated sodium channels (Nav). However, whether intracellular [Na+] is also involved in mammalian organ regeneration is unknown, because of lacking suitable mammalian models. Deer antlers are a novel model for studying mammalian organ morphogenesis. It is now well known that antlers generate from antlerogenic periosteum (AP) and regenerate from pedicle (antecedent of antler) periosteum (PP). Cells from the AP and PP have stem cell attributes, thus termed antler stem cells. In the present study, the intracellular sodium ion of the cells from the AP, PP and facial periosteum (FP) were strained in vitro with CoroNa Green indicator dye.
Expression of voltage-gated sodium channel (Nav) genes was detected using RT-PCR. Results showed that intracellular [Na+] in three cell types was significantly different: AP cells>PP cells>FP cells. Expression of the currently known ten Nav genes were all detected in the AP cells; whereas, expression of Nav1.1 gene not detected neither in the PP nor the FP cells, and expression of Nav 1.1 and Nav 1.4 genes not detected in the FP cells. Difference in Nav gene expression in the different antler stem cells may contribute to the different intracellular [Na+] in these cells, which then may correlate with the degree of the stemness of antler stem cells. We think our new finding may open up a new avenue for the study of the relationship between intracellular ion concentration and stemness of stem cells in general, hence the stem cell-based organ regeneration in mammals.
Keywords: Deer Antler; Stem Cell; Generation and Regeneration; Intracellular Sodium; Nav Gene
Abbrevations: Nav: Voltage-Gated Sodium Channels; AP: Antlerogenic Periosteum; PP: Pedicle Periosteum; FP: Facial Periosteum; [Na+]: Sodium Ion Concentration
Recently, level of intracellular sodium ion concentration ([Na+]) has been found as a functional regulator of organ generation and regeneration [1]. Voltage-gated sodium channels (Nav) play an important role in controlling and maintaining the balance of intracellular and extracellular sodium ions [2]. Thus far, 10 types of Nav genes have been identified (Nav1.1 ~ Nav1.9 and NavX genes) and knockdown of Nav1.6 had significant negative effects on zebrafish motoneuron development [3]. Early increase in intracellular [Na+] is required for initiating regeneration following Xenopus tail amputation, while molecular and small chemical inhibition of influx of sodium ion in blastema causes failure in regeneration [1]. Ectopic eyes were reportedly induced through microinjection of neonatal Nav1.5 channel mRNA into all four cells at the four-cell stage or through controlling of influx of sodium ions when incubated in sodium gluconate in Xenopus laevis [4].
However, current knowledges with regard to intracellular sodium transportation are solely based on lower form animals, such as amphibians. It is not clear as to whether these findings can be applied to the situation of mammals, as thus far mammalian models are lacking. Deer antlers are a novel model for studying mammalian organ morphogenesis [5]. It is a unique mammalian organ that can fully grow back after lost, which offers a rare opportunity to explore the instructive signals for organ generation and regeneration in mammals [6]. Deer antlers generate from antlerogenic periosteum (AP, the periosteum overlying a deer frontal crest) and regenerate from pedicle periosteum (PP; the periosteum enveloping a permanent bony protuberance, known as pedicle) [7-9]. It is known that the PP cells are the direct derivative of the AP cells [10].
Cells from the AP and PP are termed antler stem cells, as they express both mesenchymal, such as CD90, CD73 and CD105 etc (Wang et al, in submission) and embryonic stem cell markers, such as CD9, Oct4 and Nanog; and have multiple differentiation potentials, such as into chondrocytes, adipocytes, myotubes and neuronal- like cells [11]. In one of our previous studies, we found that development of deer antlers was effectively impaired by the treatment with MS-222 (a chemical that can effectively inhibit influx of sodium ion) on antler stem cells in vivo [12]. Therefore, intracellular [Na+] and Nav gene expression must have implicated in antler development. In the present study, intracellular [Na+] and Nav gene expression status of the AP and PP cells were investigated, and we found that intracellular [Na+] was positively associated with the degree of stemness of antler stem cells. Our results provide the first evidence that intracellular [Na+] and expression of its regulator Nav genes may be involved in deer antler generation and regeneration, the reminiscent in lower level animals.
The AP, PP and deer facial periosteum (FP) were collected following the protocols as previously described from 2 male sika deer heads immediately after slaughtering [13-15]. Collection of the tissue samples was approved by the local Animal Ethics Committee (CAAS2017046C). Cells released from these tissue types were cultured in the culture medium (DMEM containing 10% FBS, 100U/ml penicillin, and100μg/ml streptomycin) at 37 °C in a 5% CO2 incubator; trypsinzed when cells reached around 85% confluence and stored in the freezing medium (FBS+10% DMSO) in liquid nitrogen at passage 3. When needed, the antler stem cells were retrieved from the storage.
The AP, PP, and FP cells were seeded at a density of 5.0×104 cells/well in 6-well plates containing 2 ml culture medium/well until reaching about 70% confluence. Culture medium was then replaced by 1 ml HBSS equilibrium solution containing 8.5 μM CoroNa (Invitrogen, C36676) for 30 min at 37 °C and 5% CO2. Fluorescent intensity of different antler stem cells was examined under EVOS microscope (Invitrogen).
When the AP, PP, and FP cells reached 90% confluence in 10-cm-dishes respectively, they were collected, and their RNA was extracted respectively with PureLink® RNA Mini Kit following the protocols provided by the manufacturer. Ten pairs of primers were designed based on the bovine Nav1.1 ~ Nav1.9 and NavX gene sequences (the NCBI GenBank). Identification of different types of Nav genes were carried out using gradient PCR.
Intracellular [Na+] was reflected by the fluorescent intensity after CoroNa staining. The order of fluorescent intensities was as follows: AP cells>PP cells>FP cells (Figure 1).
Figure 1: Fluorescent intensities of the AP (A), PP (B) and FP (C) cells after CoroNa staining. Note that the AP cells had the highest intensity, and then followed by the PP and FP cells. As fluorescent intensity reflects the intracellular [Na+] and the PP cells are the cells that are further differentiated from the AP cells, the intracellular [Na+] might be positively associated with the degree of stemness of different antler stem cells. Bar =200μm.
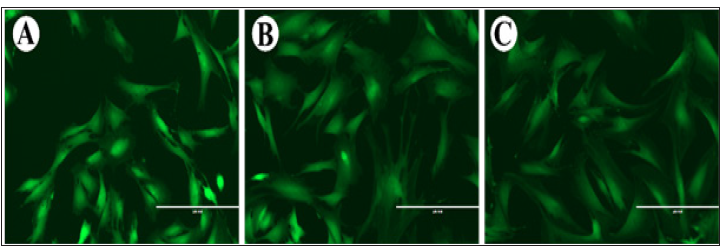
Figure 2: PCR results for Nav genes of 1.7, 1.4, 1.1
M: marker; 1: Nav1.7 gene; 2: Nav1.4 gene; 3: Nav1.1 gene.
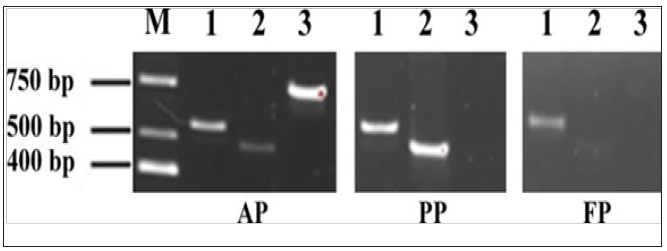
The results showed that all currently known 10 Nav genes were detected in the AP cells at the transcriptional level; whereas, only 9 Nav genes detected in the PP cells (Nav1.1 gene missing), and 8 Nav genes detected in the FP cells (both Nav1.1 and Nav1.4 genes missing) (Figure 2). Therefore, the higher fluorescent intensity, thus the higher intracellular [Na+], in the AP cells may be because the expression of Nav1.1 gene; the higher fluorescent intensity, thus the higher intracellular [Na+], in the PP cells may be because the expression of Nav1.4 gene.
The results showed that all currently known 10 Nav genes were detected in the AP cells at the transcriptional level; whereas, only 9 Nav genes detected in the PP cells (Nav1.1 gene missing), and 8 Nav genes detected in the FP cells (both Nav1.1 and Nav1.4 genes missing) (Figure 2). Therefore, the higher fluorescent intensity, thus the higher intracellular [Na+], in the AP cells may be because the expression of Nav1.1 gene; the higher fluorescent intensity, thus the higher intracellular [Na+], in the PP cells may be because the expression of Nav1.4 gene.
It is reported that terminally differentiated cells tend to be strongly polarized, while more plastic cell types (stem cells, embryonic cells, and cancer cells) tend to be relatively depolarized [18]. As is known, Nav genes play an important role in maintenance of the cell membrane potential; a small but significant number of sodium ions moving into the cells depolarizes them [19]. Further study is needed to determine if the AP cells is more depolarized than the PP cells. Our finding in the present study that there exits positive association between intracellular [Na+] and the degree of stemness of antler stem cells would open a new avenue for the exploration of stem cell-based mammalian organ regeneration, the Holi grail in the regenerative medicine.


