Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Andrea Maria Guarino, Alessandra Pollice and Viola Calabro*
Received: May 31, 2018; Published: June 20, 2018
*Corresponding author: Viola Calabro, Dipartimentodi Biologia, Italy
DOI: 10.26717/BJSTR.2018.05.001258
Using fluorescent proteins as imaging probes is a wide spread and versatile technique in microscopy. GFP tagged proteins can be used to track and examine real time localization, interactions and translocation of proteins of interest as well as to investigate several aspects of cell biology. The Y-box binding protein 1 (YB-1) is a pleiotropic protein involved in a widenumber of cellular processes and a substantial amount of knowledge on YB-1 localization and function was produced using YB-1-GFP constructs. Recently, it has been shown that YB-1 plays a critical role in cell stress response, playing an important role in the formation of stress granules (SG). To deeply investigate on this subject, we produced a stable HEK293T cell line constitutively expressing YB-1-GFP. In physiological conditions YB-1-GFP expressing cells behaved like the parental cells and YB-1-GFP protein properly localized to the cytoplasmic compartment. However, upon oxidative insult we observed a strong reduction in cell viability along with the occurrence of unusual GFP positive aggregates. Taken together, our findings suggest tocritically revise existing insights obtained with YB-1-GFP construct and, morein general, to beware and be critical in interpreting data obtained withfunctionally uncharacterized GFP fusion proteins.
Keywords: GFP; Stress Granules; Oxidative Stress; Stable Cell Line
Abbrevations: CSD: Cold Shock Domain; NES: Specific Nuclear Export; NLS: Nuclear Localization; CRS: Cytoplasmic Retention Signals; SG: Stress Granules; RNP: Ribonucleoproteins; CLS: Cell Line Service; mRNPs: Messenger Ribonucleoproteins
Y-box binding protein1 (YB-1) is a member of the evolutionarily conserved cold shock domain (CSD) proteins and was first identified as a DNA/RNA binding protein [1] involved in the control of gene expression at both transcriptional and translational level [2-4]. Given its multiple cellular functions, YB-1 is involved in the control of severalbiological processes including cell proliferation and migration. To properly perform its functions, YB-1 subcellular localization has to be finely regulated. Specific nuclear export (NES), nuclear localization (NLS) and cytoplasmic retention signals (CRS) contribute and direct the multifunctional tasking of YB-1 [5]. In normal resting cells, YB-1 localizes to cytoplasm where it is a majorcomponent of P-bodies and messenger ribonucleoproteins (mRNPs) [6]. Recent studies link YB-1 tothe cellular response tooxidative stress and DNA repair mechanisms. Indeed, following acute oxidative stress, YB-1 localizes to cytoplasmic Stress Granules (SGs), organelle-likestructures devoid of membranes engagedin mRNA sorting and pro-survival translational reprogramming [7,8]. In particular, YB-1 is recruited in TIA-containing Stress Granules (SGs) where it functions as a component of translationally inactive mRNPs todirectly block translational initiation of highly expressed [9]. Following DNA damage, YB-1 translocates to the nucleus andassociates with DNA repair protein complexes. Here we report the characterization of a stable pool of HEK293T cellsconstitutively ex pressing YB-1 as a GFP fusion protein enabling sensitiveanalysis of YB-subcellular localization by confocal microscopy.
Cell Culture: HEK293T, human embryonic kidney cells were purchased from Cell LineService (CLS, Germany). Cells were cultured in humidified incubator at 37°C and 5% CO2 and maintained in DMEM High Glucose (Gibco) supplemented with10% Fetal Bovine Serum (Gibco), 1% L-Glutammine (Gibco) and 1% Pen-Strepsolution (Gibco). For HEK293T YB-1-GFP and HEK293T GFP, blasticidin( Sigma-Aldrich, St Louis, MO) and G418(Sigma-Aldrich, St Louis, MO)antibiotics were added in the medium to a final concentration of 5μg/ml.
Plasmids: The expression construct pcDNA6V5/HIS-A-YB-1- GFP derived from pcDNATM3.1(+) (Invitrogen); pcDNA 3.1HIS-C was purchased from Thermo Fischer Scientific (USA). peGFP-C1 was purchased from Clontech(USA).
Transfections: Cells were transfected using Lipofectamine 2000 (Life Technology) according to the manufacturer’s instructions. Briefly, cells were seeded at 70-80% confluence (1.5 × 106) in 100mm dishes and transiently transfected with different concentrations of plasmid (from 800ng upto 1,5μg).
Generation of YB1-GFP Stable Clones: HEK293T cells were tested for their sensitivity to blasticidin in order to determine the best concentration for a correct selection. Concentrations of 2,5, 8, 10 and 12mg/ml were tested. Controls start to die from the day after the addition of the higher blasticidin concentration up to 3 days for the lower antibiotic concentration. The concentration chosen for selection was 5mg/ml. HEK293T cells were seeded in 100mm dishes (Corning) and let grow to reach 80% confluence. Cells were transfected by lipofectamine with pcDNA6/V5-HISA encoding human YB-1-GFP bearing the gene for blasticidin resistance or with pcDNA3.1/His C control plasmid. After 48 hours from transfection, blasticidin was added to plates at a final concentration of 5mg/ml. Aspreviously observed, pcDNA3.1/His C transfected cells start to die after 3 daysof selection. After one week of selection cells were tripsynized and replated and after two weeks of selection stable clones were obtained
Immunofluorescence Microscopy: 3x104 HEK293T, HEK293T GFP and HEK293T YB-1-GFP cells were seeded onpoly-D-lysine pre-treated coverslips (12mm thickness); the day after, cells were treated or not with H2O2 (Sigma-Aldrich, St Louis, MO). At the end of treatment cells were gently rinsed with 1X PBS before adding PFA at 3,4% for 10 minutes to fix cells; after 3 washes with 1X PBS cells were permeabilized by adding 0.5% Triton X-100-PBS for five minutes; then, after 3 washes with 1X PBS, cells were blocked to avoid unspecific binding of antibodies with a 3% BSA solution for 20 minutes. Primary antibodies were added for 1hour (dilution range1:200); after incubation, three washes with 0.05% Tween PBS were performed; then Alexa Fluor 488 (Life Technologies) and Cy3 (Jackson Immuno Research, USA) secondary antibodies were added for 45 minutes in dark; after 3 washes in 0.05% Tween PBS cover- slips were immersed in a glycerol PBS solution and fixed. Direct eGFP signal was acquired without the addiction of fluorescent dyes. Images were acquired using a Carl Zeiss LSM700 equipped with an axio Observer Z1 or with a Nikon TE Eclipse 2000. Image processing and analysis were performed with Fiji (ImageJ) software.
Cell Viability Assay: Cell viability was determined by the MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay (Sigma-Aldrich, St Louis, MO) as previously described [10]. Cells were seeded in 96-well plate at the cell density of 2 x103; the day after cells were treated or not with H2O2. After treatment, cell medium was substituted with DMEM without phenol-red (GIBCO) and without supplements; MTT solution was added to cells to produce formazan crystals. After 2hrs MTT solution was substituted by acidic isopropanol to solubilize theformazan crystals. The optical absorbancewas determined at 570nm using an iMark microplate reader (Bio-Rad, USA). The experiments were carriedout intriplicate for each experimental point and reported as percentage of the untreated control set at 1.0.
Trypan Blue Assay: Cells were seeded on poly-D-lysine pre-treated 12 well plates at 2,5 x105. The day after plating, cells were treated or not; after treatment cells weregently rinsed with 1X PBS, trypsinized andcollected. An aliquot was diluted 1:1 with trypan blue (Sigma-Aldrich, St Louis, MO). The experiments were carried out in triplicate for each experimental point.
Cell Proliferation Analysis: Cells were seeded on poly-D-lysine pre-treated 12-well plates at 2,5 x105. Every 24 hours cells were gently rinsed with 1X PBS, trypsinized and counted in a Burker chamber. The count was confirmed with analysis by a Scepter 2.0(Millipore). The experiments were carried out in triplicate for each experimental point.
Immunoblot Analysis: Cells were seeded at 60% confluence (1.5 × 106) in 100mm plates; the day after, plates were gently rinsed with 1X PBS, and collected in Lysis Buffer (50mM Tris-HCl pH 7.5, 5mM EDTA, 150mMNaCl, 1% NP-40, 1Mm phenylmethylsulphonyl fluoride, 0.5% sodium deoxycholate, and protease inhibitors). Cells were then left on ice for 30 minutes, and crude extracts were obtained following centrifugation at 13200rpm for 30 minutes at 4°C. Theamount of protein in the samples was determined by the Bio-Rad protein assay (Bio-Rad, Milan, Italy). After the addition of Laemmli buffer (Sigma Chemical Co, St. Louis, MO, USA) the samples were boiled at 100°C for 5min and resolved by SDS- polyacrylamide gel electrophoresis (SDS-PAGE). The proteins werethen transferred to a polyvinylidene difluoride membrane (PDVF, Millipore) using a Mini trans-blot apparatus (Bio-Rad, Milan, Italy) according to the manufacturer’s instructions. The PVDF membrane was blocked in 5%w/v milkbuffer (5%w/v non-fat dried milk, 50mMTris, 200mM NaCl, 0.2% Tween 20) and incubated with primary antibodies diluted in 5% w/v milk or bovine serum albumin (BSA) buffer overnight at 4°C. The blots, washed three timeswith TTBS (Tris-bufferedsaline, 0.1%Tween), were incubated for 1 hour atRT with HRP-conjugated secondary antibodies (SigmaAldrich). Proteins were visualized by an enhanced chemi luminescence method (ECL, GE-Healthcare) and analyzed by Quantity One W software of ChemiDoc TM XRS system (Bio-Rad, Milan, Italy).
Primary Antibodies: Anti-YB-1 (Abcam, UK); anti-PABP (Sigma- Aldrich, St Louis, MO); anti-GAPDH(Santa Cruz Biotechnology, USA) (Table 1).
Note: Statistical analyses were performed using GraphPad Prism7 (GraphPad Software Inc). Statistical significance of difference in measured variables between control and treated groups was determined by the two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons post-test. Difference were considered significant at P< 0.033 (*), P< 0.002 (**) and P< 0.001 (***).
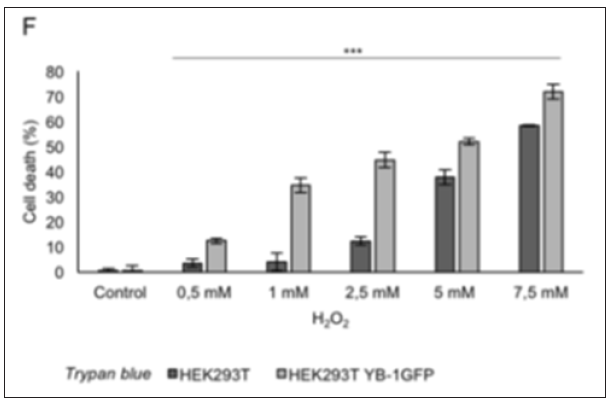
Stable cell lines represent a permanent resource that can be stored under cryogenic conditions for long periods of time, retrieved, and cultured to provide a consistent and reliable level of sustained gene expression. To explore the role of YB-1 localization upon stress stimuli we sought to generate a stable pool of cells constitutively expressing YB-1as a GFP fusion protein. Preliminarily, we transiently transfected HEK293T cells with YB-1-GFP cDNA cloned in pcDNA6/V5-His, a vector derived from pcDNATM3.1(+) (Invitrogen). 5.0x105 cells were seeded in order to reach 80% confluence the day after; lipofection was performed using pcDNA6/V5- HisA-YB-1GFP and peGFP-C1 empty vector as control [10]. 48hours after transfection cells were analyzed by immuno blot to evaluate the level of YB-1-GFP fusion GFP exogenous expression. Transfected YB-1-GFP was highly expressed inHEK293T cells (Figure 1). Direct fluorescence, acquired by Nikon Eclipse 800 microscope, showed a prevalent cytoplasmic localization for YB1-GFP (Figure 2). Moreover, viability of YB-1-GFP cells, as assessed by MTT assay, was undistinguishable from HEK293T parental cells and cells expressing GFP alone (data not shown). Next, we produced pools of clones of HEK293T stably expressing YB-1-GFP or GFP alone. YB-1- GFP expressing cells were selected by blasticid in while G418 was used for selection of peGFP-C1 stable pool. After 4 weeks, 91% of selected cells were GFP positive. Next, we analyzed the proliferation rate of YB-1-GFP expressing pool. Compared to the parental cell line, YB-1-GFP expressing cells grew slightly faster (Figure 3). To examine YB-1 protein localization under oxidative stress we treated HEK293T YB-1-GFP cells with hydrogen peroxide (H2O2). Byimmunofluorescence, we could observe typical stress granules (SGs) in control and GFP expressing cells, using a canonical SGs marker, PABP-1 Figure 4 left panel. Unexpectedly, YB-1 fusion protein was detected in unusual GFP positive aggregates that were ab normal in both dimension and shape (Figure5 right panel arrows). Remarkably, although YB-1-GFP cells grew faster than control cells, their sensitivity to oxidative stress was increased as demonstrated by MTT and try pan blue assays (Figure 6). Taken together, our data show that, even though the GFP fusion protein behaved like the endogenous one and is localized in the right sub cellular compartment, the presence of the huge GFP tag causes a lack in functionality that can be observed only in particular experimental conditions. Therefore, insights obtained so far with YB-1-GFP construct should be critically revised and, more in general, we should beware and be careful in interpreting data obtained with functionally uncharacterized GFP fusion proteins.
Figure 1: Western blot of total extracts from HEK293T YB-1-GFP and control (untransfected) HEK293T cells, showing the full length endogenous 50 kDa YB-1 b and and the YB-1-GFP bands (66-90 kDa); GAPDH was used as loading control
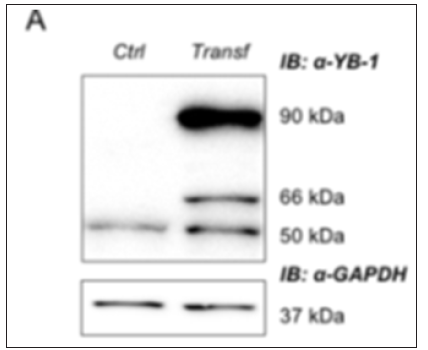
Figure 2: Direct eGFP fluorescence of HEK293T YB-1-GFP and HEK293T GFP in physiological conditions; images were acquired using a Nikon TE Eclipse 2000 (20 x objective).
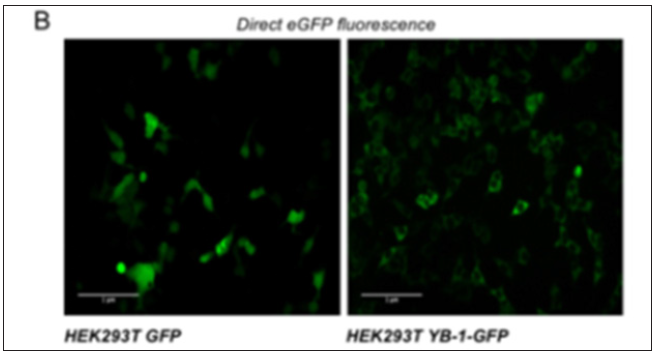
Figure 3: Cell proliferation profile of HEK293T and HEK293T YB-1-GFP from time 0 (seeding time) to 96 hours (2-way ANOVA P<0,001***, for post-hoc tests see Table 1).
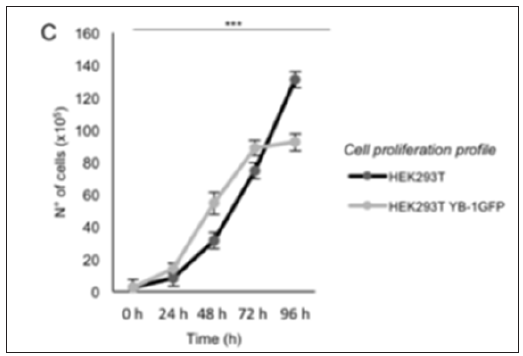
Figure 4: (left panel) confocal immunofluorescence of HEK293T treated with 0,5mM hydrogen peroxide (H2O2) for 1 hour; PABP (red) and YB-1 (green) mark Stress Granules; (right panel) direct eGFP fluorescence of HEK293T, HEK293T GFP and HEK293T YB-1GFP treated with 0,5mM hydrogen peroxide (H2O2) for 1 hour; images acquired using a Carl Zeiss LSM 700 (40x oil objective) (left panel) and Nikon TE Eclipse 2000 (20 x objective) (right panel).
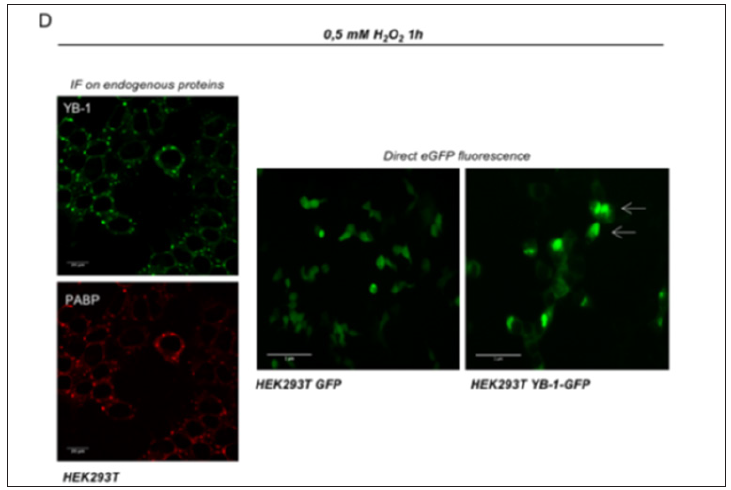
Figure 5: cell viability assay (MTT) of HEK293T and HEK293T YB-1-GFP cells treated with indicated concentrations of hydrogen peroxide (H2O2) for 1 hour (2-way ANOVA P<0,001***, for post-hoc tests see Table 1).
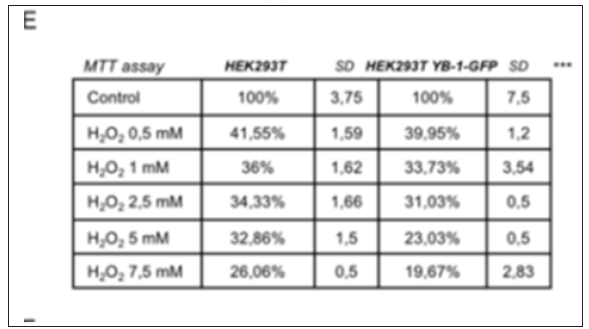
Figure 6: Trypan blue count; representation of the increasing number of dead cells after hydrogen peroxide (H2O2) treatments.