Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Andrew B Bonner*
Received: May 22, 2018; Published: May 30, 2018
*Corresponding author: Andrew B Bonner, Auburn University College of Veterinary Medicine, Scott-Ritchey Research Center Auburn, Alabama, USA
DOI: 10.26717/BJSTR.2018.05.001142
Clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR associated protein 9 (Cas9) form a complex known as CRISPR-Cas9, which is an efficient tool for gene editing that was discovered through the study of bacterial defense mechanisms against foreign nucleic acids. This technology allows for the knock-out or knock-in of specific DNA sequences. Therefore, CRISPR-Cas9 may provide a new method to abrogate the effects of genes that lead to disease or introduce genes that promote improvements in health. The powerful nature of these potential uses can be illustrated by considering the genetic bases of diseases such as cancer or obesity. Clearly, there will be many ethical debates about the future of this technology to alter genetic function.
Abbreviations: CRISPR: Clustered Regularly Interspaced Short Palindromic Repeat, PAM: Protospacer Adjacent Motif, HR: Homologous Recombination, NHEJ: Non-Homologous End Joining, TALENS: Transcription Activator-Like Effector Nucleases, EGFP: Enhanced Green Fluorescence protein
The ability to manipulate the genome is a fairly new field of biology that was brought about by the discovery of recombinant DNA technologies in the 1970’s. The field has continued to progress through the use of novel techniques into a new field known as genome engineering [1-3]. Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (CRISPR-Cas9) is an innovative technology that has been revolutionizing the field of biology and more specifically the field of genome engineering. This system functions as an accurately targeting restriction endonuclease that is guided by a hybrid strand of RNA. The hybrid strand of RNA consists of a strand known as the crRNA (or CRISPR RNA), which is approximately 20 bp in length, and a transactivating CRISPR RNA (tracrRNA) [1]. The hybrid RNA strand is known as a single guide (sgRNA) strand. This RNA strand forms a complex with a Cas-9 endonuclease.
The complex is maintained through interactions between the tracrRNA strand and the Cas-9 endonuclease [1,2]. The crRNA is used to create a complementary base pair strand with a target strand of DNA [2]. This crRNA binds with the target genome sequence through normal Watson and Crick base pairing. These interactions are depicted in (Figure 1). The crRNA sequence is variable and can be programmed to be complementary with a target sequence [3]. This interchangeability of the crRNA sequence makes the CRISPR- Cas9 complex very accurate in targeting specific sequences. This specificity has made CRISPR technology revolutionary in the category of restriction endonucleases [3]. The technology has a basis as a microbial defense mechanism, and this review will discuss the discovery of this system in these microbial systems and future directions for this technology as the limits of its usage are tested.
The CRISPR-Cas9 complex was first discovered through its role as a type of bacterial and archaeal defense mechanism against foreign nucleic acids [3]. More specifically, the CRISPR-Cas9 system is used as an immune system response in these cells against viral DNA [4]. The primary mechanism of the CRISPR-Cas9 system acts similarly to immune systems in eukaryotes. When a virus infects a bacterial or archaeal cell, part of the viral DNA sequence is incorporated into the CRISPR region in the bacteria [4]. The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) region of RNA is found in most types of bacteria and archaea. For some microbial species, the CRISPR regions can account for approximately 1% of the entire genome of the organism. The abundance of these regions in many microbial regions may contribute to their ability to frequently undergo horizontal transfer into other microbial or- ganisms [4]. The locus consists of several repeated regions that are usually 21 to 48 bp in length with interspersed sequences of RNA that are often complementary to DNA from viruses [3,4].
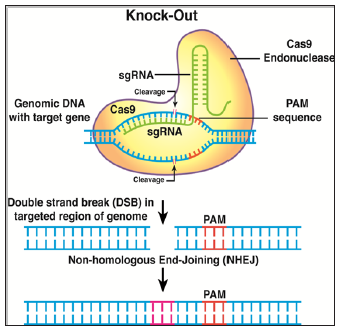
Figure 1: CRISPR/Cas9 knock-out of target gene sequences results in gene-specific disruption in host cells. The CRISPR/Cas9 complex aligns with the complementary DNA sequence in the host cells. The Cas9 endonuclease produces a site-specific DNA double strand break within the gene of interest (due to the complementary basepairing of the crRNA sequence and the target sequence) and adjacent to a PAM sequence. The DNA double strand break is repaired through error-prone non-homologous end-joining, which results in abrogation of gene function (see text).
These interspersed sequences are known as spacers and are complementary to viral protospacers. The mechanism by which spacers are added into the CRISPR is still being investigated, but it is certain that the basis for spacer development comes from viral protospacers [4]. In most cases, a protospacer adjacent motif (PAM) bearing the sequence, NGG, is present in the sequence for the protospacer that is used to generate the spacer that is eventually inserted into the CRISPR region. The presence of the PAM sequence will become more important when considering interactions with Cas proteins in a later section. When more spacers are incorporated between repeat elements in the CRISPR locus, the bacteria are able to acquire a broader spectrum of resistance against invading viral nucleic acids [3]. Located near the repeat sequences are a family of Cas genes. The Cas9 gene is particularly important for the CRISPR- Cas9 immune system as it is the gene that codes for the Cas9 restriction endonuclease [5].
The Cas9 restriction endonuclease is important for the bacterial immune response as it interacts with portions of the CRISPR locus (pre-crRNA) after they have been modified to interact with the Cas9 endonuclease [3]. The pre-crRNA (CRISPR RNA) can be prepared for interaction with the Cas9 endonuclease through a few different processes of modifications that involve cleavage of repeat regions by ribonucleases and RNases. The final crRNA product is combined with a second sequence known as tracrRNA. This association of the crRNA and tracrRNA sequences is known as an sgRNA and is combined with the Cas9 endonuclease mainly through interactions between the tracrRNA and Cas9 [3]. This complex of sgRNA and Cas9 forms into a CRISPR ribonucleoprotein complex (crRNP) [4].
Once this complex is formed, the widely known use of the CRISPR- Cas9 technology to cleave double-stranded DNA can be carried out (Figure 1). The complex is able to scan a genomic sequence in search of a complementary protospacer sequence for the crRNA sequence. The sequence that is selected, will contain a PAM sequence directly next to the complementary genomic sequence, which will provide a signal for the Cas9 endonuclease to cleave the DNA at this point [4]. A double-stranded DNA break is created that is approximately three base-pairs upstream of the PAM. Subsequently, DNA repair mechanisms are activated: non-homologous end joining (NHEJ) and homologous recombination (HR). These events can lead to knock-in or knockout of various genes [6].
The basic structure and usage of CRISPR-Cas9 is revolutionary in the field of genome editing tools. Before CRISPR, the main genome editing tools were known as transcription activator-like effector nucleases (TALENS) and Zing Finger Nucleases. These tools were fairly effective in editing target DNA sequences, but they had two major shortcomings: they were prone to cause off-target effects and they were protein-based rather than RNA-based. This latter trait made it more difficult to synthesize and incorporate the required components into cells. The CRISPR system displays improvement in these areas as CRISPR has been shown to have fewer off-target effects than other genome-editing tools, and it is RNA-based, which is easier to synthesize and incorporate into target cells [7]. Another advantage of CRISPR that has recently been discovered is the use of nanoclews as a means to effectively introduce the CRISPR-Cas9 complex into cells. Because of the size and slightly unstable nature of the CRISPR-Cas9 complex, one major setback regarding the potential therapeutic use of the CRISPR complex has been incorporating the complex into cells. Nanoclews provide a shuttle for the CRISPR complex in a unique way. Nanoclews are essentially a circular piece of yarn that is composed of DNA, and they are produced by rolling-circle replication that is common in bacterial plasmids. The nanoclews provide complementary base pairs to the sgRNA of the CRISPR complex and shuttle the complex into the cell. From there, the CRISPR complex is able to enter the nucleus and interact with the DNA of the cell [8].
Knock-out of Targeted Sequences: One logical use of CRISPR technology is to knock-out genes and observe the result of the loss of function of this gene. The mechanism that governs this process has been previously described in the description of the mechanism and functionality of the CRISPR-Cas9 complex. Figure 1 provides a visual depiction of the knock-out process using the CRISPR- Cas9 complex. As displayed in Figure 1, the knock-out process is dependent on creating a double-strand break through the use of the CRISPR-Cas9 complex that will be repaired through the error- prone process of non-homologous end-joining [9]. An example of an important CRISPR knockout study involves the regulation of Cadmium content in indica rice through the use of CRISPR technology. Due to Cadmium’s negative health benefits and its increasing presence in indica rice, one of the most common forms of rice in the world, it has become increasingly important for producers and consumers of this form of rice to lower the Cadmium levels found in this rice [10]. OsNramp5 is a protein involved in the transport and integration of metals, including Cadmium, into the root systems of rice plants. For this reason, the OsNramp5 gene has been viewed as a potential target for knock-out as a means of reducing the levels of OsNramp5 protein and eventually Cadmium levels in the rice. Therefore, investigators targeted an exon of the OsNramp5 gene through the CRISPR-Cas9 complex to create a mutation in the gene. This mutation in the OsNramp5 gene resulted in an extremely significant decrease in the Cadmium levels of the indica rice that was tested [10]. These results provide promising potential for the use of CRISPR technology to create healthier foods that can benefit the health of populations around the world.
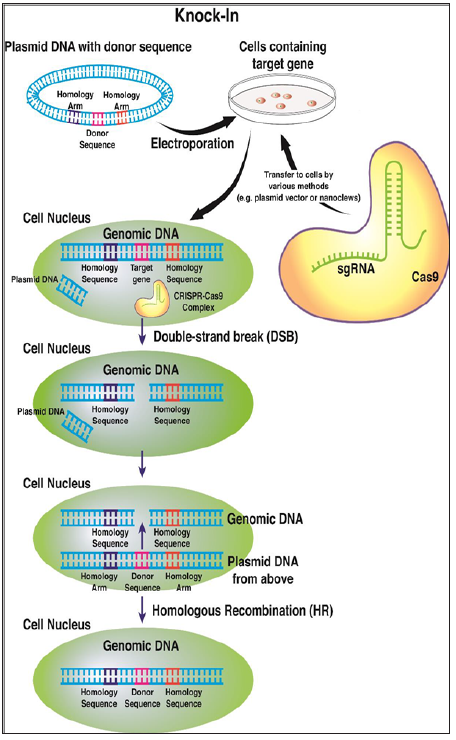
Knock-in of Donor Sequences: One of the major possible uses of CRISPR technology involves using CRISPR to incorporate foreign DNA into genomic DNA. The process of creating a knock-in mutation has been well-documented, and current studies are attempting to optimize the knock-in process. The first important step for beginning a knock-in experiment using CRISPR technology is the identification of a safe harbor locus. These loci are regions of the genome that are especially able to accommodate insertion of foreign nucleic acid based on favorable conditions such as 3D arrangement of the surrounding genomic DNA. One of the most common safe harbor loci is the Rosa26 locus, which is located on chromosome [11,12]. Following the identification of the safe harbor locus, the next step is to develop two plasmid vectors that will be inserted into a cell line that contains genomic DNA with the target gene of interest. The first plasmid vector contains sequences that will express a site-specific sgRNA sequence and the Cas9 protein. As described earlier, the sgRNA sequence will be complementary to the target gene of interest, and it will form the complete CRISPR-Cas9 complex with the Cas9 protein. Other forms of delivery of the CRISPR-Cas9 complex into the cell are possible, and the most efficient process for delivery of the complex is the subject of investigation and may depend on the gene of interest [13-16]. The second plasmid contains the donor sequence of interest flanked on either side by homology arms that correspond to the sequences flanking the target gene of interest [12] (Figure 2). The CRISPR-Cas9 complex will form a double-strand break in the gene of interest, and this break will be repaired through homologous recombination by the second plasmid containing the homology arms. Thus, a new sequence will be introduced in the place of the target gene of interest [12]. Current studies are underway to optimize this knock-in process using CRISPR technology [13-16]. One study used similar parameters to those described above, but several of the parameters were slightly modified in attempts to optimize the frequency of successful knock-in of the donor DNA. The study measured several different characteristics of the knock-in experiment that could potentially optimize the knock-in process using CRISPR technology. Initially, the investigators sought to optimize the conditions of electroporation using various voltage settings and cell lines in order to insert a plasmid containing a donor sequence into cells [12]. The next parameter that was tested was the selection of the most efficient sgRNA for creating site-specific double-strand-breaks. Several criteria were taken into account in order to select the most effective sgRNA. They found that it was very important to select an sgRNA that is most likely to bind to the target region and not bind to any off-target regions. Using these criteria as a guide, it was found that the most effective sgRNA sequence was able to create a mutation efficiency of 34.3% [12]. The final parameter that was optimized was the length of the homology arms (Figure 2) used in the donor plasmid to aid in the process of homologous recombination in order to insert the donor sequence into the genomic DNA. The study found that the longest homology arm tested (1.5 kb) was the most effective at creating a site-specific insertion of donor DNA at the site of the double-strand break [12]. This finding indicates that increasing the length of the homology arms may increase the likelihood of insertion of a donor sequence into genomic DNA. Many additional studies will continue to investigate the optimization of these CRISPR processes with the aim of maximizing the effectiveness of knock- in sequences.
Figure 2: CRISPR/Cas9-directed knock-in of donor gene sequences is a precise technique to modify gene expression in host cells. The donor sequence that has been cloned into a plasmid vector is electroporated into host cells at the safe harbor sites. The CRISPR/Cas9 complex is delivered intracellularly through various techniques (see text). The interaction of the CRISPR/Cas9 complex and the genomic DNA results in a site-specific DNA double strand break. Through interaction of homologous sequences of genomic DNA and plasmid DNA, the donor sequence is inserted into the genomic DNA by the process of homologous recombination.
Cancer as an Example for Potential CRISPR-Cas9-Directed Therapy: Cancer is one of the most harmful and widespread diseases in the world currently. The genetic basis of cancer has become an enormous area of scientific exploration. Genetic causes of cancer can be extremely complex, and many research efforts to this date, have provided very few possible therapeutic solutions to these genetic abnormalities [7]. The introduction of CRISPR- Cas9 technology has provided a new tool to use in the struggle for researchers to uncover the complex genetic basis of different types of cancer. Cancer researchers are attempting to use CRISPR-Cas9 technology to identify target genes for drug development. The CRISPR mechanism is able to accomplish this goal by creating libraries of potential cancer-related genes through knocking out these genes and observing the effect of the loss of function of these genes [7]. This method of gene identification not only applies to cancer genes, but almost any disease- causing gene in the genome. Many laboratories have begun to create models for different types of in vivo cancer-related studies using CRISPR technology. The CRISPR technology is able to provide a loss of function phenotype for cancers that are known to have a genetic basis [7]. These findings are important for future investigation as they provide a way to view the entire process of disease development from the genetic basis to the phenotype of the disease [7]. An example of a possible cancer-related target for CRISPR/Cas9 is the tumor suppressor gene TP53 [17]. This tumor suppressor gene is mutated in approximately 50% of cancers. The mutations in this gene can take place throughout its sequence, but the mutations are commonly missense mutations that lead to impaired function of the TP53 protein. Under non-mutated conditions, the TP53 protein is involved in apoptosis and senescence of tumor cells. Loss of these events leads to more aggressive growth of tumors and a poor prognosis. Investigators have proposed using CRISPR genome editing technology to replace mutant TP53 with wild type TP53. Some investigators have proposed delivering a CRISPR-based construct through a viral vector that is encapsulated in a tumor-targeted phage nanoparticle. The goal of these efforts is to knockout mutant TP53 and replace it with functional TP53 [17]. Therefore, these latter studies may provide methods for correcting genetic defects that would lead to cancer-causing genes or to reinstitute genetic function that is related to protection of cells from malignant transformation. Furthermore, these techniques may allow scientists to alter genetic expression in cancer cells and thereby render cancer cells more susceptible to cancer treatments.
Obesity as an Example for Potential CRISPR-Cas9-Directed Therapy: CRISPR has great therapeutic potential for many deadly diseases, but it also has potential to play a role in preventing health conditions that predispose patients to disease. Obesity is becoming an increasingly prominent problem in America, and it can be the basis for other diseases such as cancer or heart disease. CRISPR technology may provide a genetic basis for obesity therapy that could lead to decreased genetic-linked obesity before it leads to more serious disease [18]. Expression of Leptin has been associated with a protective effect on obesity [18]. One particular study used CRISPR technology to target the Leptin gene in rats in order to assess its effects on obesity as well as other possible food intake problems that may come with loss of function of the leptin protein. The Leptin gene was chosen for this study because of its known relation to the regulation of food intake and energy usage. The study found that knockout of the Leptin gene greatly increased the rates of obesity in rats. This increase in obesity rate was directly related to the loss of function of the Leptin protein [18]. Therefore, in the future it may be possible to upregulate Leptin function and potentially decrease the debilitating effects of genetic-linked obesity.
Ethical Usage of CRISPR Technology: Since its discovery, many people have feared the ethical dilemmas posed by the possible clinical implementation of CRISPR technology. The fears are so great that in 2015, leading scientists in the field of biology called for a worldwide moratorium on the use of CRISPR technology to edit the human germ line [19,20]. Scientists who are involved in genetic research have various viewpoints on this issue. Some feel that it is the place of researchers to uncover more therapeutic uses for CRISPR technology as its role in disease prevention could be a great benefit to society. Other scientists are wary of the possible misuse of CRISPR technology as the powerful nature of the technology has already been described, and the accessibility of the technology is rapidly increasing [19,20]. Both of these viewpoints hold merit, and the debate is still continuing.
The debate has been intensified because of the rapid pace at which CRISPR technology has progressed [18-20]. Decisions on future areas of research will be forthcoming. The potential for this technology to be used in utero on unborn fetuses will be topic for much debate in the future. Genetic editing with CRISPR technology has recently taken on new ramifications. Investigators have shown that this technology can be used to knock-in or knock out-out genes in utero [9,10,21-23]. This manipulation of genes in utero may have far reaching effects on genes that may change the health and well-being of unborn offspring. For instance, recent studies have shown that electroporation technology can be used to knock-in neural progenitor genes such as the mouse βIII-tubulin (Tubb3) gene, and the effectiveness of this knock-in can be assessed by the Enhanced Green Fluorescence protein (EGFP) in utero in mammalian models of mice and ferrets [21].
These investigators demonstrated a high efficiency of in utero knock-in that has significant implications for the future of this technology. If genes can be consistently placed in humans in utero by similar technology, the potential of this technology seems limitless. One might ask, can we add genes that augment intellectual integrity or athletic prowess? These are important questions that will need to be addressed further as this technology advances. Additionally, investigators have explored the use of this technology to delete genes that may lead to suboptimal survival or early fatality at the in utero stage. Shinmyo et al. have explored electroporation and its potential to knock out brain-specific genes in utero [9]. They used CRISPR technology to target the Satb2 gene which is responsible for abnormalities in axonal projection processes. They showed that knock out of this gene was consistent with the role of Satb2 in the process of cell-autonomously developing callosal axon projections [9]. Again, the implications of these studies are paramount, as in utero knock out of mammalian genes, especially neuronal genes, raises significant possibilities for the correction of brain defects in utero.
However, the ethical considerations of these endeavors have been much analyzed in the scientific literature and other editorials. Clearly, science has advanced into a new arena with the potential of CRISPR gene editing technology. These gene editing technologies are truly an exciting scientific advance. We may be able to treat diseases that were once thought to be untreatable. However, we may be able to alter genes that fundamentally change the characteristics of individuals that are the very essence of their beings. This later fact has wide- spread ethical implications for the future. Should we knock-out cancer-causing, obesity-causing or dementia-causing genes in utero, or should there be restraints on these efforts until we decide the ultimate ramifications of gene-editing? Eventually, investigators will be challenged by these questions. We are entering a new phase of genetic manipulation that may impact the substance of individuals and this great responsibility will require much attention by the scientific community of the future.
The meteoric rise of CRISPR technology, which is arguably the most important scientific discovery of this decade displays the importance of microbiological and genetic research. The fact that this technology, that is shaping the discussion of genetic engineering and medicine, is derived from a seemingly small research endeavor into bacterial behavior, demonstrates the powerful nature of biological research. At this point in time, the possible effects of CRISPR technology on the future of society are fairly unlimited depending on how society plans to use this technology in the future. Ethical debates will continue to intensify as knowledge of this technology and its limits become further known. These ethical debates will grow as CRISPR technology becomes more applicable to altering the genetic makeup of embryos [24]. These potential alterations of genetic makeup will create new avenues to correct diseases but may lead to genetic engineering that requires much consternation and debate.
The development of CRISPR/Cas9 methodology for gene editing has obviously created much enthusiasm due to the potential to treat common diseases as noted above. The power of this technology has led authors to liken these techniques to “Jedi against the dark empire of disease” [25] and propose the use of “CRISPR to build a more powerful CAR” [26].The latter reference to CAR was made regarding CAR T cells and the use of CRISPR/Cas9 technology to insert CAR cDNA at different locations in order to better control its expression with different promoters [27]. The above comparisons of CRISPR/Cas9 to superheroes or powerful vehicles demonstrates the current enthusiasm and wide-spread research into this new mode of genetic manipulation.
This work was performed as part of an honors microbiology project. The author would like to thank Dr. Yohannes Mehari (Auburn University) for advice and Dr. Robert Kesterson and Dr. Anil Challa (University of Alabama at Birmingham, School of Medicine) for mentorship regarding the use of CRISPR-Cas9.


