Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Lei Zhang1, Lihong Weng2 and Robert J Hickey*3
Received: May 25, 2018; Published: May 29, 2018
*Corresponding author: Dr Robert J Hickey, Associate Professor of Molecular Medicine, Beckman Research Institute of the City of Hope National Comprehensive Cancer Center, 1500 E Duarte Road, Duarte, CA 91010, USA
DOI: 10.26717/BJSTR.2018.05.001131
The resolution of alkaline proteins by 2D-polyacrylamide gel electrophoresis (2D-PAGE) is often accompanied by protein streak artifacts. To overcome this type of artifact formed during traditional 2D-PAGE, and to further improve the resolution of alkaline proteins, we developed a DeStreak ™ protocol that essentially eliminates the streaking artifact through the use of hydroethyldisulfide (HED) throughout the entire IEF run. We found that HED improves the separation and resolution of individual basic proteins when using a narrow range (pH 7-10) IPG strip format.
Keywords: 2D-polyacrylamide gel electrophoresis, Isoelectric Focusing, De-streaking
Abbrevations: HED: Hydroxy Ethyl Disulfide, 2-VP: 2-Vinyl Pyridine
One of the challenges of using 2D-polyacrylamide gel electrophoresis (2D-PAGE) for the analysis of the proteome is the inherent difficulty associated with resolving basic proteins [1]. Horizontal streaking within the alkaline region of 2D-PAGE gels is frequently observed. Two of the major reasons for this streaking are the loss of reducing agent in the basic part of the IPG strip, which is used for the first dimension separation, and re-oxidation of thiol groups on the proteins. Protein aggregation through the formation of intra- and inter- protein disulfide bridges then cause this streaking. At alkaline pH, the commonly used reducing agent Dithiothreitol (DTT) becomes negatively charged, and dissociates from these alkaline proteins; where it then migrates toward the anode, which subsequently depletes it from the basic region of the gel. The use of Tributylphosphine (TBP) is a more effective alternative to DTT because TBP remains unchanged over a broad pH range. TBP, however, has a somewhat limited solubility, and is unstable in solution. It is also prone to loss during the first dimension separation. Streaking can also be minimized by alkylation of the sulfydryl groups of cystein and reduction of the disulfide bridges of the protein. Iodo acetamide (IAA) and 2-Vinyl pyridine (2-VP) are commonly used alkylating reagents for this purpose. Recently an equilibrium reac tion alternative to alkylation, Hydroxy Ethyl Disulfide (HED), was found to protect the thiol groups by transforming disulfide bridges into highly stable disulfides, to yield a single protein species [2]. In this study, the high efficiency and effectiveness of the De Streak™ approach using HED were observed and are recommended for use with proteins having an alkaline isoelectric point (pI).
Nuclear Extracts were prepared from MCF-7 cell pellets according to the TECHNICAL BULLETIN offered with the CelLytic™ Nu- CLEAR™ Extraction Kit (Sigma-Aldrich, Sigma-NXTRACT). Protein concentration was determined using a Thermo Scientific Pierce 660 nm protein assay (Cat: #22662). 50 μg of Nuclear Extract was loaded onto a 7 cm/pH 7-10/IPG Strip using anodic cup loading for the first dimension, followed by resolution of these proteins using a 10% SDS-PAGE slab gel for the second dimension. The standard rehydration protocol (7M Urea, 2M Thiourea, 4% CHAPS, 1% DTT) and the DeStreak™ protocol (7M Urea, 2M Thiourea, 4% CHAPS,1.2% HED, GE Healthcare, Cat: # 71-50025-42 AE) were compared. The resolved proteins were then stained with Coomassie Blue and detected using the GS-800™ Calibrated Densitometer with Quantity One 1-D analysis software. A Modified Bio-Rad IEF protocol was used: (i.e., “Extended 300v Protein Entry Step”). Step A: 150v, Rapid Ramp, 30 min; Step B: 300v, Gradual Ramp, 2.5-5hours; Step C: 4,000v, Gradual Ramp, 1 hour; Step D: 4,000v Rapid Ramp, 8,000 VH; Step E: 500v, Hold.
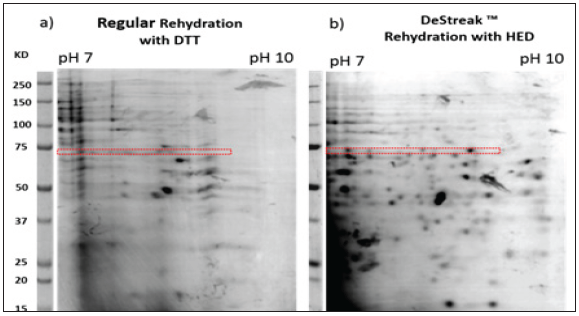
We found that the presence of HED throughout the entire IEF run greatly reduces streaking within the alkaline side of the pH7-10 IPG strip. Nuclear proteins tend to exhibit a somewhat alkaline pI. Nuclear proteins are involved in a myriad of normal and pathology driven processes, some of them may function as cancer selective biomarkers. Our work therefore, provides an effective solution for improving the analysis of nuclear protein related biomarkers of biological processes using 2D-PAGE (Figure 1).
Figure 1: The effective improvement in the resolution of individual alkaline proteins in the basic range using the DeStreak™ protocol and 2DE-IPG Strips is shown. Qualitative differences in the proteins resolved using the standard rehydration protocol with DTT (a), vs. the DeStreak™ reagent protocol (b), were compared. A representative section of horizontal de-streaking was indexed in the box. The separation of proteins is greatly improved and yields more single spots within the alkaline region.
The work described in this manuscript was supported in part by a Cancer Center Support Grant awarded by the National Institutes of Health to the City of Hope under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.


