Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
W Grant Cooper*
Received: March 27, 2018; Published: April 11, 2018
*Corresponding author: W Grant Cooper, International Physics Health and Energy, Inc., Lubbock, TX 79424, USA
DOI: 10.26717/BJSTR.2018.03.000949
Recent studies [1-4] of EPR (Einstein-Podolsky-Rosen) - generated [5] entangled proton qubit-pairs demonstrate that Grover's [6] enzyme quantum-readers quantify quantum informational content embodied within entangled proton qubits of rat and human genomic systems [7]. Double-helical DNA is replicated into a "metastable" keto-amino state; so, metastable hydrogen bonded amino (-NH2) genome protons encounter quantum uncertainty limits [1,2], | Δx Δpx ≥ h/2 . The resulting quantum mechanical proton - proton interaction introduces probabilities of EPR-arrangements, feto-amz'no-(entanglement) → enol-imine, yielding "qubit-pairs" of entangled enol and imine protons (Figures 1 & 2). Reactions yielding EPR-generated entangled proton qubit-pairs are observed [1,2] as G-c → G'-C',G-C →*G-*C and A - T→ *A - *T A. (Bold italics - G'-C',*G - *C,*A - *T - denote necessity of Hilbert space to describe dynamics of embedded entangled proton qubit-pairs; see Figures 1 & 2 for notation).
Figure 1: Schematic of "metastable" keto-amino and "ground state" enol-imine hydrogen bonds.
Amino protons encounter quantum uncertainty limits, ΔxΔpx ≥ h /2 , introducing probabilities of EPR arrangements, keto- amino-(entanglement) → enol-imine. The asymmetric double-well potential represents an energy surface "seen by" metastable hydrogen bonding amino proton, and a "ground state", entangled enol and imine proton "qubit-pair". Product enol and imine protons are entangled, and are each shared between two indistinguishable sets of electron lone-pairs, and thus, participate in entangled quantum oscillations, +;>⇄/ -> , between near symmetric energy wells, occupying intramolecular decoherence-free subspaces.)
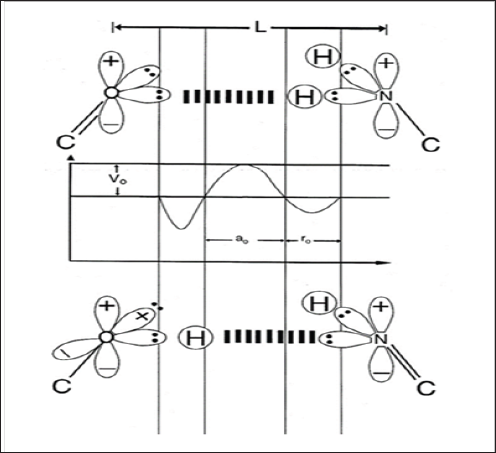
Figure 2: Distribution of entangled proton qubit states at a G'-C' (symmetric) or *G-*C (asymmetric) superposition site. Symmetric, asymmetric and second asymmetric (unlabeled) channels (→) by which metastable keto amino G C protons populate enol and imine entangled proton qubit states. Dashed arrows identify pathways for quantum oscillation of enol and imine proton qubits. Approximate electronic structures for hydrogen bond end groups and corresponding proton positions are shown for the metastable keto amino duplex (a) and for enol and imine entangled proton qubit states, G'-C' (b-e). Electron lone pairs are represented by double dots, :, and a proton by a circled H. Proton states are specified by a compact notation, using letters G, C, A, T for DNA bases with 2's and 0's identifying electron lone-pairs and protons, respectively, donated to the hydrogen bond by - from left to right - the 6 carbon side chain, the ring nitrogen and the 2 carbon side chain. Superscripts identify the component at the outside position (in major and minor groves) as either an amino group proton, designated by 00, or a keto group electron lone pair, indicated by 22. Superscripts are suppressed for enol and imine groups.)

Dynamic, EPR-generated entangled proton qubit-pairs, |+> ⇄ |->, occupy "intra-atomic", decoherence-free subspaces (Figure 1) of enol oxygen and imine nitrogen [1-2], and thus, are resistant to decoherence by "normal" in vivo classical environments [24]. Grover's [6] enzyme quantum reader "measures", δt << 10-13 s, quantum informational content by "trapping” an entangled proton qubit-in ket-state, within a genome groove [1-2]. These measurements, >□t << 10-13 s, provide instructions for executing quantum information processing, | Δt≤≤ 10-14 s [1-4], of entangled proton qubits, which specify evolutionary distributions of the 22 most abundant rat and human microsatellites [2-4,7]. Since the evolutionary "emergence-distance” between rat and human is ~ 70x106 y [8], EPR-generated entangled proton qubit-pairs must retain entangled "two-state” quantum coherence,+> ⇄ |-> , for years to decades, before specifying microsatellite evolution instructions with "measured” quantum informational content of EPR-generated entangled proton qubit-pairs [1-4]. Otherwise, Grover’s-type [6] quantum processor measurements, δt << 10-13 s, of EPR-generated entangled proton qubit-pairs could not accurately specify evolutionary distributions of the 22 most abundant microsatellites common to rat and human [7], as exhibited [3-4].
These results [1-4,7-8] imply that EPR-generated entangled proton "qubit-pairs” [5] are resources for quantum information processing in all double helical DNA systems, including human brain-cell DNA [1-4,9]. In this case, quantum information processing, | Δt ≤ ≤ 10-14 s [1-4], of EPR-generated entangled proton qubits, executed by a single brain-cell, could communicate the resulting quantum information processing calculations to the brain’s neuronal network of ~ billons of neurons [9]. This evolutionarily coordinated "network” communication could cause coupled, entanglement-enabled information-processing computations in ~ billons of brain-cell DNA systems, and consequently, could execute the phenomenon of consciousness [9-10]. A model for Homo sapienS consciousness [2,9] is thus implied by ~ 3.6x109 y of entanglement-enabled evolution [1,3,4], from duplex RNA- ribozyme segments, into double helical DNA of ~ 6.8x 109 bp, which resides in Homo sapiens brain-cells [1-4].
Homo sapiens’ brain contains sensory "biological files" [1-4,9] for vision, acoustics, vocal, olfactory, touch, taste, balance, selfmotion, pain, emotion, language, analyses, music, imagination, "truth" and "fantasy". Sensory files are interfaced with, initially vacant, "hard drive" memory files, which are ultimately populated by "stable” EPR-generated, entangled proton qubits [1-4]. Braincell DNA of robust human infants has been evolutionarily selected to accumulate "stable” EPR-generated entangled proton qubits [14], for purposes of storing quantum information, and executing its quantum processing [1-2]. Brain-cell "DNA reservoirs” is incrementally populated with EPR-generated entangled proton qubits to their "threshold limits” [1-4], which is achieved by age ~25 y for H. sapiens. Consciousness is a consequence of Grover's [6] processors continually conveying quantum informational instructions for consciousness-relevant quantum information processing of entangled proton qubits.
Anesthetics [10] inhibit Grover's processors, which cause rapid transitions from consciousness to unconsciousness. This explanation implies "normal”, non-drug-induced sleep is a consequence of "inactive” Grover’s [6] processors, which-during consciousness-actively measure quantum informational content of EPR-generated entangled proton qubits. Thus, normal sleep is a "down-time” condition that allows EPR-generated entangled proton qubits to repopulate "recently measured” segments of double helical DNA [1-4], while Grover's processors are "inactive” In this case, the "sleeping”, unconsciousness brain is operating on "autopilot”, without the "total component” of Grover’s processors continuously "reading” quantum informational content embodied within EPR-generated entangled proton qubits, which provides instructions for consciousness-relevant quantum information processing.


